Levitra enthält Vardenafil, das eine kürzere Wirkdauer als Tadalafil hat, dafür aber schnell einsetzt. Männer, die diskret bestellen möchten, suchen häufig nach levitra kaufen ohne rezept. Dabei spielt die rechtliche Lage in der Schweiz eine wichtige Rolle.
Recent advances in bioorthogonal reactions for site-specific protein labeling and engineering


Contents lists available at
Tetrahedron Letters
advances in bioorthogonal reactions for site-specific proteinlabeling and engineering
Yukang Gong, Lifeng Pan
Key Laboratory of Bioorganic and Natural Products Chemistry, Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, Lingling Road 345, Shanghai 200032,People's Republic of China
In the past two decades, with the rapid development of chemical biology, tremendous small-molecule
Received 15 November 2014
based toolkits were created by organic chemists, and were widely used to study and manipulate proteins
Revised 11 March 2015
in order to dissect their complicated biological functions. This review summarizes some recent
Accepted 12 March 2015
progresses of bioorthogonal reactions for site-specific protein labeling and engineering, and highlights
Available online
the powers of using these methods to study the biological functions of some proteins.
Ó 2015 Published by Elsevier Ltd. This is an open access article under the CC BY-NC-ND license
Bioorthogonal organic chemistryLigand-directed organic chemistryProtein labeling and engineering
Bioorthogonal reactions with genetically encoded unnatural amino acids for site-specific protein labeling and engineering . . . . . . . 00Bioorthogonal ligation reactions . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 00
Deprotection reactions through photo-uncaging. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 00Deprotection reactions through palladium-mediated uncaging . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 00Ligand-directed reactions for site-specific protein labeling and engineering . . . . . . . . . . . . . . . . . . . . . . . 00
critical to the development of biomedical and biotechnologicalapplications. Traditional genetics, molecular biology, biochemistry,
As one of the most abundant biomolecules, proteins are
cell biology, and allied methods have provided various tools to
involved in most of the biological processes and perform a wide
investigate the functions of proteins, and have led to tremendous
array of important functions within living organisms. Therefore,
achievements including visualization of a protein using fluorescent
the study and manipulation of protein functions are not only of
protein fusions and silence of a protein expression using RNA
significant importance to fundamental scientific research, but also
interferences. However, not all the proteins and related biologicalprocesses are within the easy reach of those conventional
approaches. Fortunately, recent rapid progress in the chemical
Corresponding author.
E-mail address: (L. Pan).
biology field provides abundant new technologies for the study
0040-4039/Ó 2015 Published by Elsevier Ltd.
This is an open access article under the CC BY-NC-ND license (
Please cite this article in press as: Gong, Y.; Pan, L. Tetrahedron Lett. (2015),
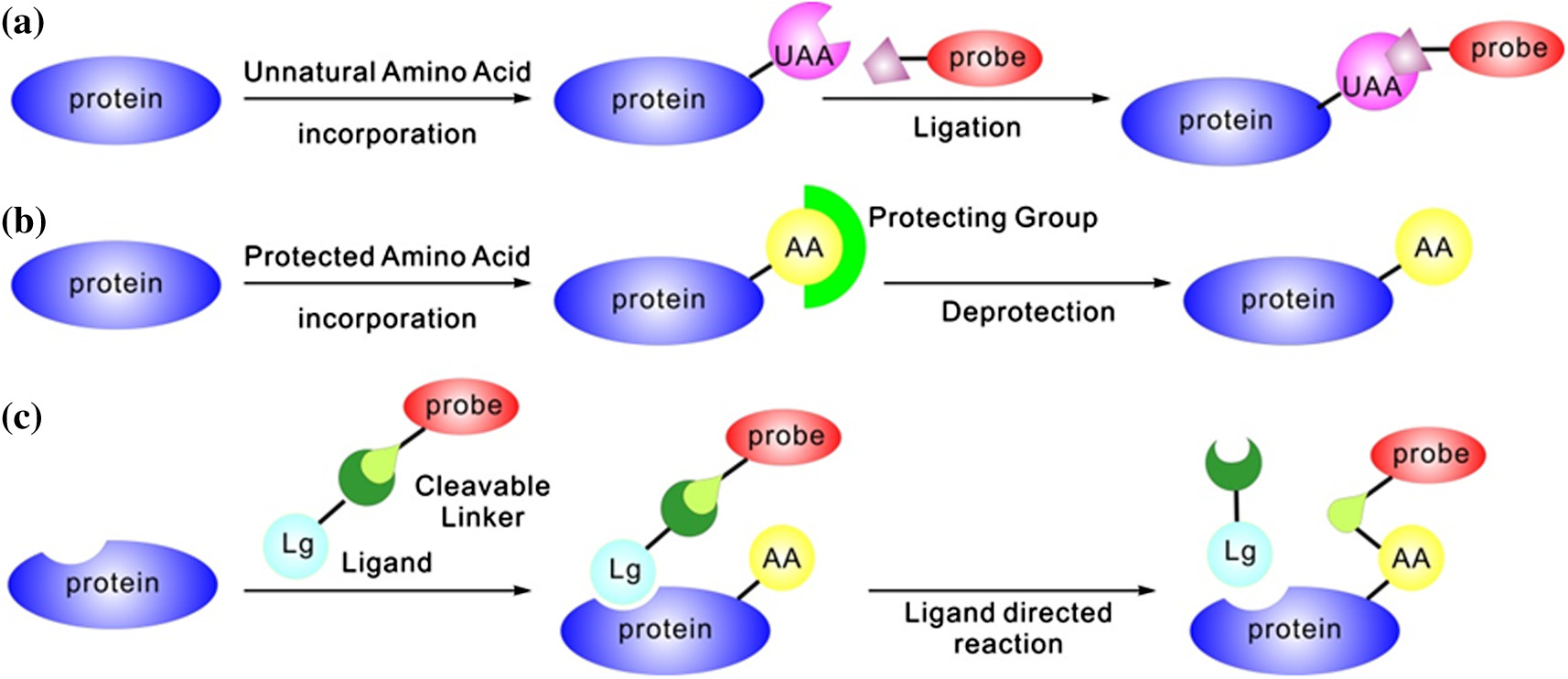
Y. Gong, L. Pan / Tetrahedron Letters xxx (2015) xxx–xxx
Figure 1. Three approaches to achieve the site-specific protein labeling and engineering. Schematic illustrations of site-specific incorporation of unnatural amino acidsfollowed by bioorthogonal reactions including ligations (a) and deprotections (b). (c) Schematic illustration of the ligand-directed chemistry for site-specific protein labelingand engineering. UAA, unnatural amino acid; AA, natural amino acid; Lg, ligand.
of these challenging proteins and cellular processes. In particular,
Since these two methods revolutionized our abilities to site-
the modification of specific proteins with functional probes
specifically label and manipulate intact proteins, these two areas
provides a powerful technique for the investigation of target pro-
are rapidly growing and many elegant applications have been
teins and their complex functions in detail. So far, there are a large
recently reported. In this review, we summarize some recent
variety of strategies developed by organic chemists to achieve site-
developments in these two fields using bioorthogonal reactions
specific labeling and engineering of target proteins with functional
for site-specific protein labeling and engineering.
small Due to space limitations, this digest only focuseson two strategies used for the site-specific protein labeling and
al reactions with genetically encoded unnatural
engineering: (1) Bioorthogonal reactions with genetically encoded
amino acids for site-specific protein labeling and engineering
unnatural amino acids bearing functional groups that can bespecifically ligated or deprotected a and b); (2) Ligand-
In general, the bioorthogonal reactions used for site-specific
directed bioorthogonal reactions for site-specific modifications of
protein labeling and engineering based on genetically encoded
target proteins (
unnatural amino acids can be mainly classified into two categories:
first strategy combines bioorthogonal reactions with
ligation reactions (a) and deprotection reactions (b).
genetically encoded unnatural amino acids bearing functional
al ligation reactions
groups, such as aldehydes, ketones, azides, and alkenes, to facilitatethe site-specific protein labeling and engineering (a and
reactions through aldehydes and ketones
Genetic code expansion and reprogramming enable the site-speci-
Genetically encoded aldehydes and ketones can specifically
fic incorporations of diverse designed unnatural amino acids into
react with hydrazides and alkoxyamines to produce stable hydra-
proteins.By evolving orthogonal ribosomes, developing mutually
zone and oxime, respectively (and were successfully
orthogonal synthetase/tRNA pairs and manipulating genomes, the
applied for the site-specific in vitro or cell surface protein labeling.
efficiency of unnatural amino acids incorporations and the num-
However the unfavorable acidic conditions and slow kinetics of
bers of unnatural amino acids that can be site-specifically encoded
these reactions prevent their applications in most intracellular
are constantly increasing. Notablely, the development and applica-
tion of the pyrrolysyl-transfer RNA (tRNA) synthetase/tRNA pair for
identified and used as a nucleophilic catalyst for both specific
unnatural amino acids incorporation have moved genetic code
cell surface and intracellular protein labeling b).
expansion from bacteria to eukaryotic cells and multicellular
Based on the classic Pictet–Spengler reaction between aldehydes
and tryptamine nucleophiles, recently Bertozzi and co-workers
genetic code expansion is a powerful tool, it also has
reported the Pictet–Spengler ligation reaction (c).In
some limitations, for example, genetic modification and subse-
this reaction, aldehydes react with alkoxyamines to form inter-
quent overexpression of proteins may perturb the physiological
mediate oxyiminium ions, which then undergo intramolecular C–
condition of cells. Thus, another strategy named ligand-directed
C bond formations with indole nucleophiles to form hydrolytically
chemistry for site-specific protein labeling that modifies selective
stable oxacarboline products. In conjunction with techniques for
endogenous proteins under their physiological conditions, was
genetic incorporations of unnatural amino acids bearing aldehydes,
developed (In this approach, a synthetic molecule con-
the Pictet–Spengler ligation provides a unique tool to generate
taining three functional groups including a target protein binding
stable bioconjugates for biomedical applications.
ligand, a reactive linker and a functional probe is constructed,and firstly its ligand part specifically binds to its target protein,
reactions through azides
then driving a bioorthogonal reaction between the reactive linker
Genetically encoded azide groups were firstly developed for the
group with an amino acid located at the vicinity of the ligand-
site-specific biomolecule labeling by the Bertozzi group through a
binding site of the target protein facilitated by the proximity effect,
process known as Staudinger ligation, a modification of the classic
finally end with the labeling of the target protein with the
Staudinger reduction of azides with triphenylphIn this
functional probe. Therefore, this method can satisfy the require-
process, the proteins bearing alkyl azides undergo ligation reac-
ments of target selectivity and site specificity.
tions to form stable amide bonds with triarylphosphine derivatives
Please cite this article in press as: Gong, Y.; Pan, L. Tetrahedron Lett. (2015),
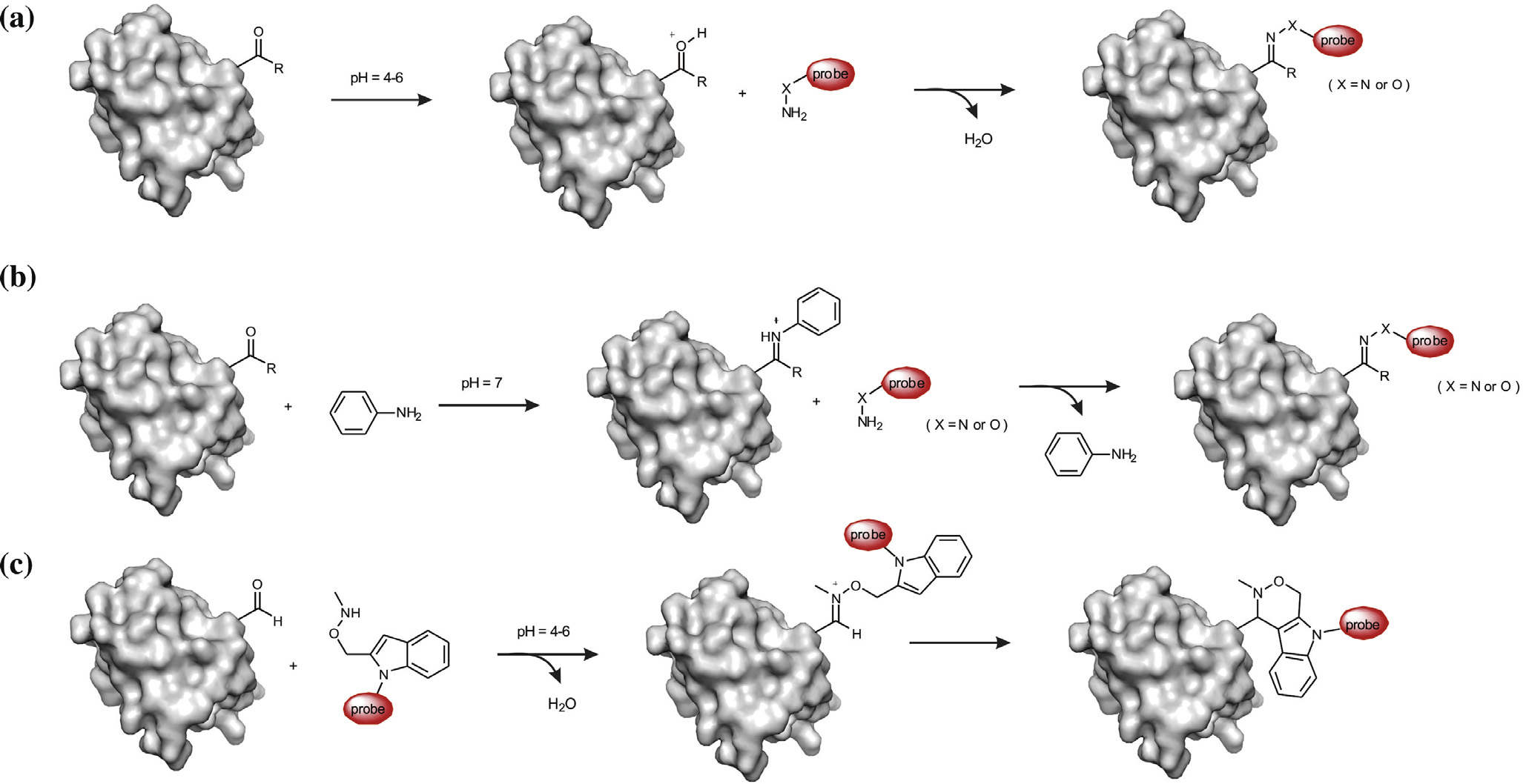
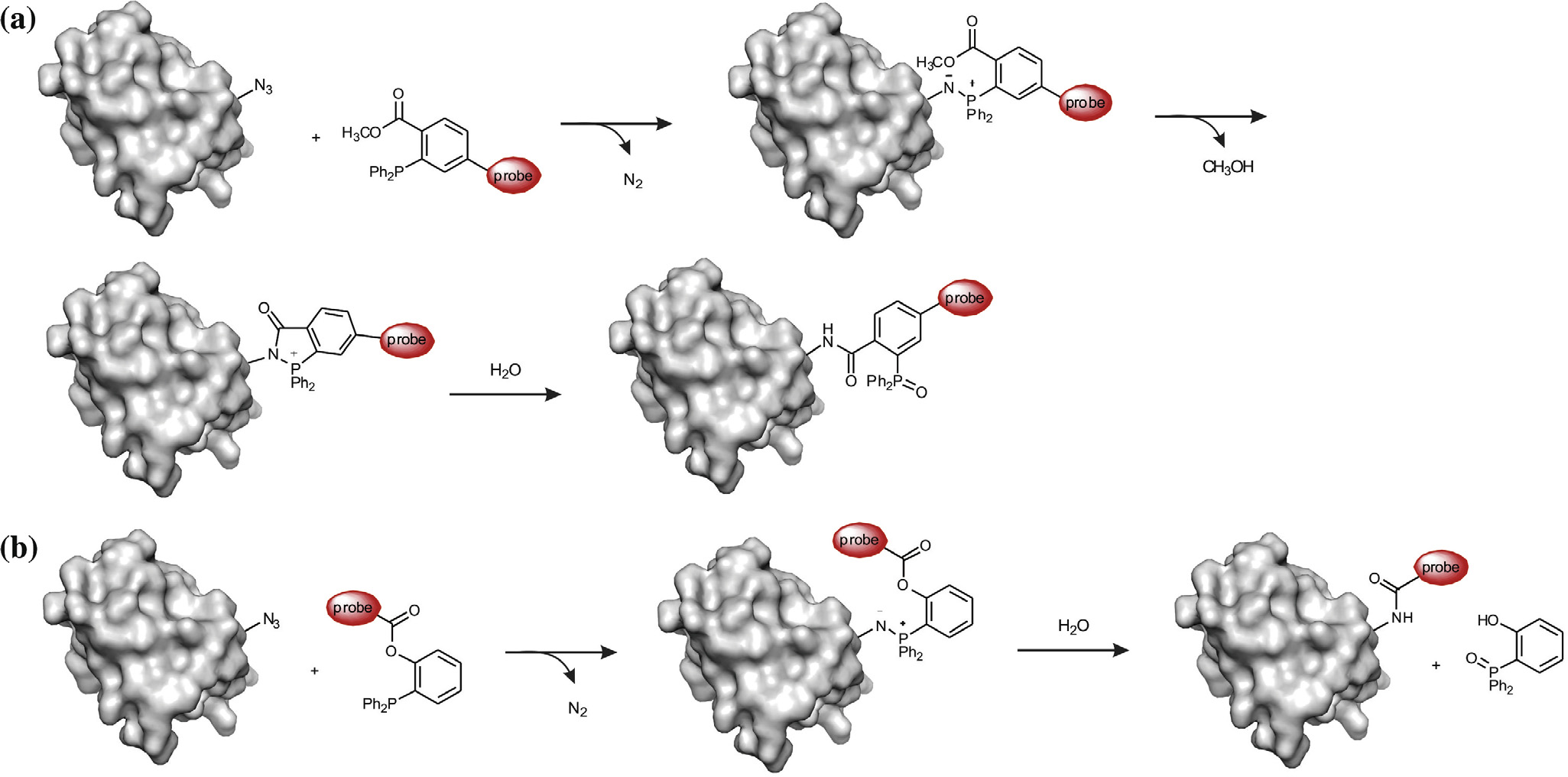
Y. Gong, L. Pan / Tetrahedron Letters xxx (2015) xxx–xxx
Scheme 1. Bioorthogonal reactions of genetically encoded ketones/aldehydes with hydrazines or alkoxyamines for site-specific protein labeling and engineering. (a) Acid-catalyzed reaction of genetically encoded aldehydes/ketones with amino nucleophiles. (b) Aniline-catalyzed reaction of genetically encoded aldehydes/ketones withhydrazines or alkoxyamines. (c) Pictet–Spengler ligation of genetically encoded aldehydes with tryptamine nucleophiles.
Scheme 2. Staudinger ligations of azides and triarylphosphines for site-specific protein labeling and engineering. (a) The Staudinger ligation between genetically encodedazides and triarylphosphines. (b) The traceless Staudinger ligation.
that have ester groups on their aromatic rings a). Later,
the Staudinger ligations are their slow kinetics and the oxidation
some phosphine reagents, in which the acyl group is attached via
sensitivities of phosphines, therefore the phosphine reagents have
a cleavable linker to the phosphine group, were developed, and a
to be used at relatively high concentrations.
variant of this reaction was reported, named as ‘‘traceless
Staudinger ligation, azides can also take part in [3+2]
Staudinger ligation'', where the phosphine oxide moiety is absent
cycloadditions with alkynes to yield stable triazoles, but high
in the final bioconjugate b).These Staudinger ligations
temperature and pressure are normally required to form the tria-
have been successfully used to site-specifically label protein
zole products in a reasonable yield.There are generally two
in vitro and in many different cellular Limitations of
strategies to improve the reaction: catalyzing the reaction with
Please cite this article in press as: Gong, Y.; Pan, L. Tetrahedron Lett. (2015),
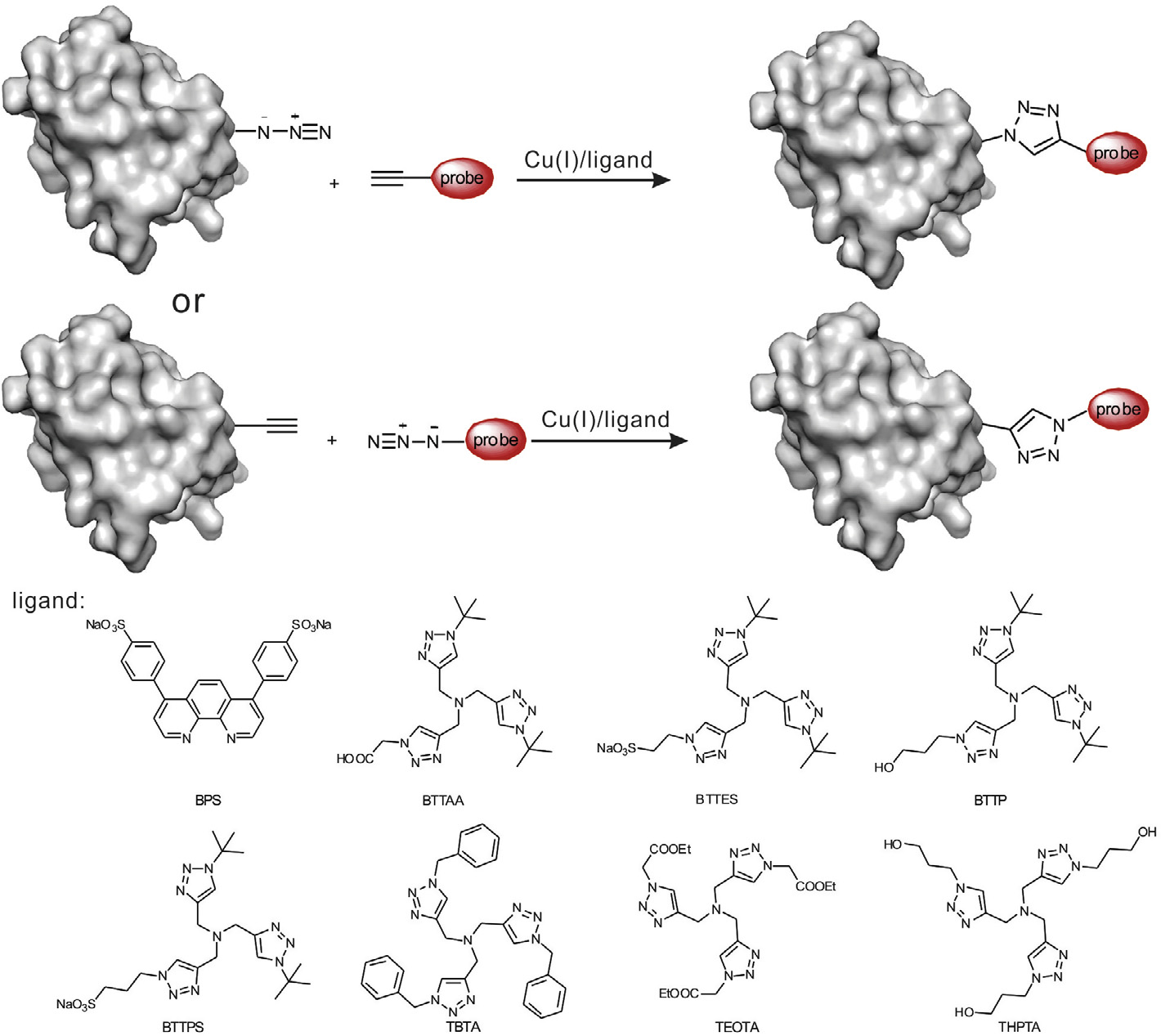
Y. Gong, L. Pan / Tetrahedron Letters xxx (2015) xxx–xxx
Scheme 3. Copper-catalyzed azide–alkyne cycloadditions (CuACC) for site-specific protein labeling and engineering together with ligands including BPS, TBTA, TEOTA,THPTA, BTTAA, BTTES, BTTP, and BTTPS for the coordination of Cu(I).
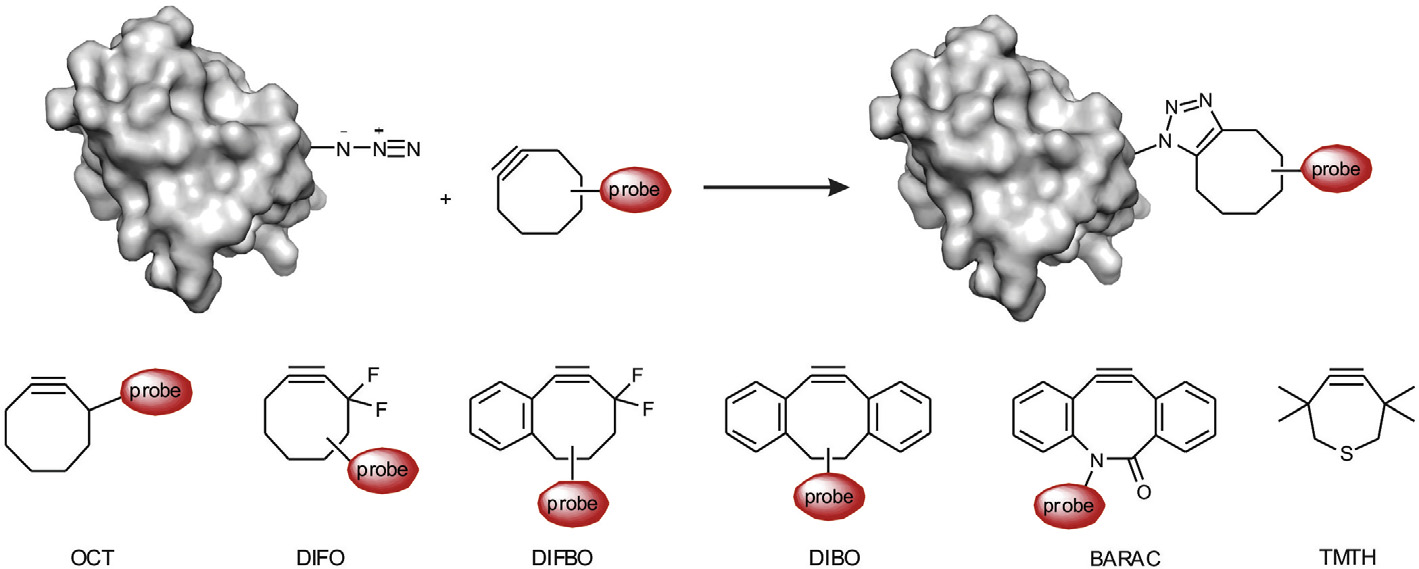
Scheme 4. Strain-promoted azide–alkyne 1,3-dipolar cycloadditions (SPAAC) for site-specific protein labeling and engineering together with strained cycloalkyne derivativesOCT, DIFO, DIFBO, DIBO, BARAC, and TMTH used in SPAAC.
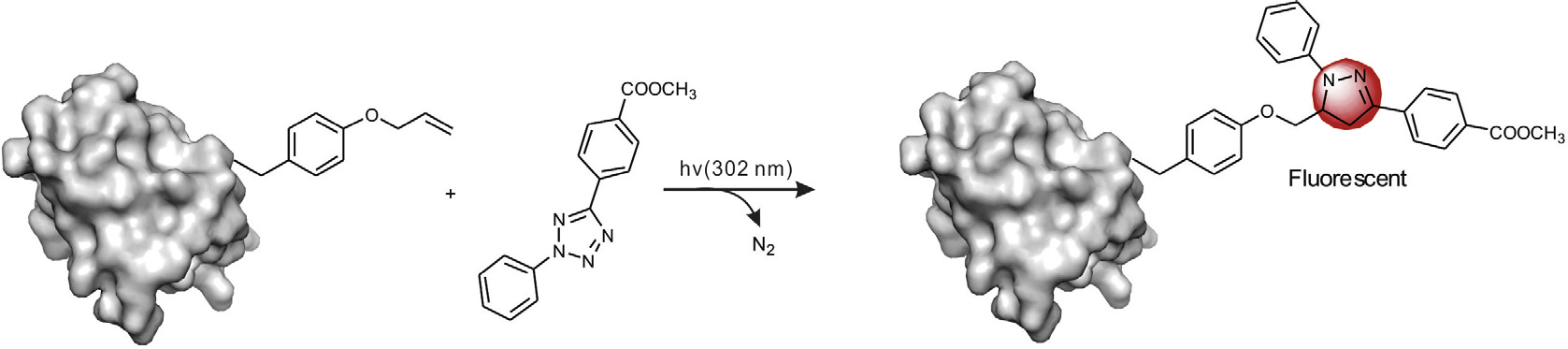
Scheme 5. Photoclick chemistry of genetically encoded O-allyltyrosines with tetrazoles for site-specific protein labeling and engineering.
copper (I)or introducing ring strain into the alkyne.The former
CuAAC, taking advantage of the formation of a dinuclear copper
is termed as the copper catalyzed azide–alkyne cycloaddition
intermediate to activate both terminal alkynes and is a
(CuAAC), and the latter is named as the strain-promoted alkyne–
representative of click reaction (Moreover, the
azide cycloaddition (SPAAC).
Cu(I)-catalyzed cycloaddition can be further accelerated by the
Please cite this article in press as: Gong, Y.; Pan, L. Tetrahedron Lett. (2015),
Y. Gong, L. Pan / Tetrahedron Letters xxx (2015) xxx–xxx
use of specific ligands for These ligands, such as BPS
Ligation reactions through strained alkenes/alkynes and
) and natural amino acid histidine, coordinate Cu(I) to
form activated copper catalysts that can promote the azide–alkyne
2008, Fox and co-worker firstly reported a protein
cycloaddition at low concentrations of Cu(I). However, the main
bioconjugation strategy based on the inverse-electron-demand
limitation of CuAAC is the toxicity of which is caused by
Diels–Alder reaction between a tetrazine compound and a strained
Cu(I)-mediated generation of reactive oxygen species (ROS) from
trans-cyclooctene.Later, various unnatural amino acids bearing
O2.Recently, the use of water-soluble ligands including TBTA,
strained alkene or alkyne groups, such as norbornenes (Nor), cyclo-
TEOTA, THPTA, BTTAA, BTTES, BTTP, and BTTPS can reduce the
propenes (Cyp), bicyclononynes (BCN), and trans-cyclooctenes
apparent copper-associated cytotoxicity by serving as reductants
(TCO or sTCO), were synthesized and genetically incorporated into
to protect cells from ROS By using these rate-
proteins for site-specific protein labeling and engineering with
accelerating ligands, the CuAAC have been used to site-specifically
tetrazines in living bacteria and mammalian cells (
label proteins in living Escherichia. coli and mammalian cells.
These reactions are very specific, extraordinarily fast, and many
SPAAC is another approach to accelerate the azide–alkyne
of the red tetrazine fluorophores, whose fluorescences are initially
cycloaddition ). Cyclooctyne derivatives have been used
quenched by the tetrazine, become strongly fluorescent upon
to label proteins containing genetically encoded azides in living
cycloadditions with strained alkenes or alkynes, making them par-
To improve the reaction, more reactive cyclooctyne
ticularly useful for the labeling of proteins with low cellular abun-
compounds including difluorocyclooctyne (DIFO) derivatives,
dances and for tracking fast protein-involved biological processes.
difluorobenzocyclooctyne (DIFBO), dibenzocyclooctynes (DIBO),and biarylazacyclooctynone compounds (BARAC) () were
Ligation reactions through cross-metathesis
developed and have been used to probe azide-containing proteins
bioorthogonal reaction involving alkenes is olefin
within complex biological systems including mammalian cells,
metathesis, one of the most powerful organic reactions for the
Caenorhabditis elegans and zebrafish embryos.Recently, thi-
construction of new carbon–carbon bonds. In 2008, Davis and
acycloheptynes, such as TMTH ), have been developed
co-workers reported the first utility of cross-metathesis in site-
as a new type of reagents for
specifically labeling proteins containing an allyl sulfide groupin vitro (They screened a small panel of alkenes and
reactions through terminal alkenes and tetrazoles
identified S-allylcysteine as the most efficient substrate for the
Photoinduced organic reactions are also explored to enable the
cross-metathesis reaction with allyl alcohol using the Hoveyda–
site-specific protein labeling processes in biological settings. Lin
Grubbs second-generation catalyst.Recently, they further
and co-workers developed the UV-light induced 1,3-dipolar
developed a rapid and efficient cross-metathesis reaction using
cycloaddition reactions between tetrazole derivativess and term-
Se-allyl-selenocysteine for in vitro site-specific labeling and engi-
inal alkenes ), also named as photoclick chemistry.
neering of proteins.
By incorporations of tetrazole and alkene groups into proteins inthe forms of unnatural amino acids, the photoclick chemistry has
Ligation reactions through palladium-catalyzed cross-coupling
been used to site-specifically label and engineer proteins in vitro,
yzed cross-coupling reactions have also been
and also to visualize proteins in living bacteria and mammalian
exploited to site-specifically label and engineer proteins. Initial
cells.There are several advantages of the photoclick chemistry:
reports using palladium-mediated cross-coupling reactions on
first and foremost, its inducibility by light makes it a powerful tool
proteins containing genetically encoded p-iodophenylalanine or
for spatiotemporal initiation of labeling reactions in living sys-
p-boronophenylalanine suffered from very low reaction conver-
tems; second, the reaction is fluorogenic and only the resulting
sions or harsh reaction Later, Davis and co-workers
pyrazoline product is fluorescent, which is helpful for living cell
developed a water-soluble palladium catalyst, a sodium salt of 2-
imaging third, the fast reaction kinetics, presenting a
significantly faster bioorthogonal reaction than the Staudinger
ligation and the Development of new tetrazole
reaction between a genetically encoded p-iodobenzyl group and
reagents, which are highly reactive and can be light-activated at
various aryl and alkenyl boronic acids a).The
wavelengths that are harmless to living cells, will make this
improved palladium-mediated Suzuki–Miyaura cross-coupling
reaction more attractive.
reactions typically reached completion within 1 h at 37 °C, and
Scheme 6. Inverse-electron-demand Diels–Alder reactions between tetrazines and genetically encoded strained alkenes or alkynes including cyclopropenes (Cyp), alkynesnorbornenes (Nor), bicyclononynes (BCN), and trans-cyclooctenes (TCO or sTCO) for site-specific protein labeling and engineering.
Please cite this article in press as: Gong, Y.; Pan, L. Tetrahedron Lett. (2015),
Y. Gong, L. Pan / Tetrahedron Letters xxx (2015) xxx–xxx
Scheme 7. Cross-metathesis of genetically encoded allyl sulfides or allyl selenides with allyl ethers using a Hoveyda–Grubbs second-generation catalyst for site-specificprotein labeling and engineering.
Scheme 8. (a) Suzuki–Miyaura cross-coupling reactions between genetically encoded p-iodophenylalanines and aryl- or alkenyl-boronic acids mediated by the water-solublepalladium catalyst, Pd(OAc)2(ADHP)2. (b) Ligand free Sonogashira cross-coupling reactions between genetically encoded alkyne groups and iodophenyls mediated by thePdNP generated from Pd(NO3)2.
Scheme 9. Deprotection reactions of a genetically encoded photocaged serine (a) or a photocaged lysine (b) using the photo-uncaging method.
were used to site-specifically label the membrane protein OmpC
b), and proved that PdNPs generated from water soluble
on E. coli surface. Another famous cross-coupling reaction,
Pd(NO3)2 can be an efficient and biocompatible catalyst for site-
Sonogashira reaction, has also been developed as a bioorthogonal
specific labeling of alkyne-modified proteins inside living E. coli
reaction to label alkyne-encoded proteins. Qing Li and co-workers
and other Gram-negative bacterial pathogens such as Shigella
firstly reported a bioorthogonal Pd-mediated Sonogashira cross-coupling reaction using a robust aminopyrimidine–palladium(II)
al deprotection reactions
complex, which selectively modify a homopropargylglycine(HPG)-encoded ubiquitin in aqueous medium as well as in
reactions through photo-uncaging
E. coli.Recently, Chen and co-workers developed a ligand-free
The activities of some proteins can be controlled through site-
specific installations of caging groups on side chains of key
Please cite this article in press as: Gong, Y.; Pan, L. Tetrahedron Lett. (2015),
Y. Gong, L. Pan / Tetrahedron Letters xxx (2015) xxx–xxx
residues that are essential for protein function. Since light irradia-
and the probe, but also as a reactive When the LDT
tion is a relatively noninvasive method, direct caging of proteins
reagent binds to the target protein, a SN2-type reaction is triggered
via a genetically encoded unnatural amino acid bearing a
due to the proximity effect between the tosyl moiety and a nucle-
photocleavable group allows photochemically control the func-
ophilic amino acid located at the vicinity of the ligand-binding
tionality or localization of the target proteins in complex cellular
pocket of target proteins, resulting in the site-specific labeling of
In conventional photocaging strategy, a photocaged
the target protein and release of the ligand moiety. This labeling
serine or lysine residue bearing a photocleavable group was
method can be applied for the site-specific modification of a vari-
ety of endogenous proteins under intracellular environm
). This photocaged serine or lysine amino acid was
The power of LDT chemistry for characterizations of both the target
converted to wild type amino acid by irradiation with relatively
protein and the ligand-binding sites has been well demonstrat
low-energy light, and was used to photochemically control the
Moreover, combining with fluorescent protein tag technology and
function of the target protein. In 2011, Chin and co-workers used
FRET imaging, the LDT chemistry has been demonstrated to con-
this photocaging strategy to control a photocaged MEK1 kinase,
struct a natural protein as a fluorescent reporter, which can be
and demonstrated a receptor independent activation of an artificial
used to detect its molecular interactions in vitro and in living
subnetwork within the Raf/MEK/ERK pathway, which provided
new insight into adaptive feedback and the kinetics of single steps
additional ligand-directed chemoselective reac-
in the MAP kinase signaling cascadesMore recently, Deiters and
tions based on different reactive linkers have been developed.
co-workers used this strategy to control a photocaged T7 RNA
Hideaki Kakeya and co-workers reported the use of 5-sulfonyl
polymerase, and demonstrated the photocaging of a synthetic gene
tetrazole as the reactive linker to develop a ligand-directed
network in mammalian The main advantages of this
reaction for site-specific protein modifications (b).
photocaging strategy are that the unnatural amino acids are site-
They employed this method to achieve the chemical labeling of
specifically incorporated and the modified proteins are directly
the cellular receptor of the natural product, cyclosporine
generated inside the cell, eliminating the requirement for addi-
Using an acyl phenol moiety as the reactive linker, Fenical group
tional transfection or injection.
demonstrated another ligand-directed chemoselective reactionfor site-specific protein modifications which was
on reactions through palladium-mediated uncaging
successfully applied to identify the target protein of the anticancer
Even using non-phototoxic light, the poor penetration ability of
natural product, marrinopyrrole A.Based on a bifunctional O-
light hinders the further utilizations of these photo-uncaging meth-
NBD unit (NBD: nitrobenzoxadiazole), Sodeoka group reported a
ods in deep tissues or intact animals. To avoid these limitations,
simple ligand-directed chemoselective probe for site-specific pro-
Chen and co-workers recently reported the development of a
tein labeling (The O-NBD unit is non-fluorescent,
palladium-mediated chemical uncaging method to control lysine-
but can be converted into a fluorogenic amino NBD group when
dependent activation of intracellular proteins (In this
reacted with a Lys residue. After using the O-NBD unit attached
method, they firstly used a genetically encoded lysine analogue
N,N-dialkyl-2-phenylindol-3-ylglyoxylamides (PIGAs), which are
bearing a propargyl carbamate group to protect a key lysine residue
ligands of translocator protein (TSPO), they were able to visualize
of the target protein, then used biocompatible and efficient
the mitochondria expressing TSPO in living
palladium catalysts, such as allyl2Pd2Cl2, to cleave the propargyl
recently, a new type of ligand-directed chemistry, known
carbamate group of the protected lysine analogue to generate a free
as ligand-directed acyl imidazole (LDAI) chemistry, has been
lysine residue. This palladium-mediated deprotection strategy was
developed (In this LDAI labeling method, a triple
further proved to work with a range of different proteins and cell
functional LDAI reagent is used, which contains a moderately reac-
lines, and was successfully used to reveal the detail virulence
tive alkyloxyacyl imidazole linker in addition to a target protein
mechanism of a bacterial Type III effector
binding group and a functional probe. Similar to the tosyl linkerin LDT chemistry, the alkyloxyacyl imidazole linker can react with
irected reactions for site-specific protein labeling and
an accessible nucleophilic residue of target protein assisted by
ligand binding. The power of LDAI chemistry has been proved by
Unlike the incorporations of unnatural amino acids by gene
its broad applications to selective chemical labeling of various
manipulation, which made the target proteins no longer endoge-
types of membrane proteins under living cell conditions for func-
nous, the ligand-directed protein labeling method can achieve
tional studies.In addition to introducing the functional probe,
the site-specific labeling of an endogenous target protein.
the LDAI chemistry can also be applied to construct caged proteins
Recently, a ligand-directed tosyl (LDT) chemistry employing a
in a rational one-step manner, which was demonstrated by Itaru
unique LDT reagent, which consists of a target protein binding
Hamachi and his co-workers They showed that the
ligand, a reactive tosyl linker and a functional probe, has been
activity of the caged carbonic anhydrase I was almost fully sup-
developed to label natural proteins a). The tosyl linker
pressed and absolutely recovered by light irradiation under
of the LDT reagent behaves not only as a linker between the ligand
in vitro Since LDAI reagents can react with Ser, Tyr,
Scheme 10. Deprotection reactions of a genetically encoded caged lysine with the propargyl group using a palladium-mediated chemical uncaging method.
Please cite this article in press as: Gong, Y.; Pan, L. Tetrahedron Lett. (2015),
Y. Gong, L. Pan / Tetrahedron Letters xxx (2015) xxx–xxx
Endoge nous pr otein
Endoge nous pr otein
En dogenou s protein
En dogenou s protein
En dogenous protein
A ctive p rote in
Inactive pro tein
A ctive pr ote in
Scheme 11. Ligand-directed chemoselective reactions for site-specific protein labeling and engineering. Schematic illustrations of the ligand-directed tosyl (LDT) chemistry(a), the ligand-directed 5-sulfonyl tetrazole chemistry (b), the ligand-directed acyl phenol chemistry (c), the ligand-directed O-NBD chemistry (d), the ligand-directed acylimidazole (LDAI) chemistry (e), and one-step construction of a caged protein by LDAI chemistry as well as the following photo-uncaging (f).
and Lys residues, while LDT reagents are reactive with His, Tyr and
The application of catalysts has also been introduced in ligand-
directed chemistry. Hamachi group pioneered the development of
catalyst-mediated ligand-directed chemoselective reactions for
Please cite this article in press as: Gong, Y.; Pan, L. Tetrahedron Lett. (2015),
Y. Gong, L. Pan / Tetrahedron Letters xxx (2015) xxx–xxx
En dogenou s protein
tyr osyl radical tra pping agents (TRTs)
En dogenou s protein
Scheme 12. Catalytic ligand-directed reactions for site-specific protein labeling and engineering. Schematic illustrations of the AGD catalyst-mediated (a) and the SETcatalyst-mediated (b) ligand-directed site-specific protein modification methods.
site-specific protein modification using the affinity-guided N,N-
engineering of endogenous proteins. However, it is worthwhile to
dimethylaminopyridine (AGD) catalysts, which transfer acyl donor
point out that the use of organic chemistry for labeling and
probes to nucleophilic residues near the ligand-binding site of the
manipulating endogenous proteins in their native conditions is
target protein through an SN2-type reaction (a).
currently still in its infancy and none of these bioorthogonal
Recently, Hiroyuki Nakamura and co-workers developed a
reactions mentioned in this review are fully perfect inside living
ligand-directed site-specific protein modification method based
cells. Thus, future efforts are expected to improve the reaction
on local single-electron transfer (SET) catalysis b). In
biocompatibility and specificity, and to enhance the reaction
this catalyst-mediated method, a single-electron transfer between
efficiency as well as to invent new organic chemistry based tools
the ruthenium tris(2,20-bipyridyl) complex ([Ru(bpy)3]2+) and a
that can efficiently and site-specifically label and engineer natural
nearby Tyr residue of the targeted protein generates the tyrosyl
proteins in living systems.
radical that can react with tyrosyl radical trapping agents contain-ing an N0-acyl-N,N-dimethyl-1,4-phenylenediamine unit through a
catalytic oxidative radical addition reaction.
Financial support from the National Natural Science Foundation
ons and perspectives
of China (No. 31470749), a ‘Thousand Talents Program' younginvestigator award of China, a Shanghai Rising Star Scholar award
Recent progress of chemical biology has provided significant
(No. 13QA1404300), the start-up fund from State Key Laboratory of
advances in the field of selective protein modifications with func-
Bioorganic and Natural Products Chemistry and Chinese Academy
tional small molecules. Two important strategies based on the
of Sciences are gratefully appreciated.
bioorthogonal reactions for site-specific protein labeling and engi-neering are summarized in this review. The development of the
References and notes
bioorthogonal reactions coupled with the genetic-code expansion
method not only revolutionized our abilities to site-specifically
label and manipulate intact proteins, but also will have important
implications for future practical applications. Meanwhile, with the
development of new reagents, the ligand-directed reactions will
become increasingly powerful for site-specific labeling and
Please cite this article in press as: Gong, Y.; Pan, L. Tetrahedron Lett. (2015),
Y. Gong, L. Pan / Tetrahedron Letters xxx (2015) xxx–xxx
Please cite this article in press as: Gong, Y.; Pan, L. Tetrahedron Lett. (2015),
Source: http://panlf.sioc.ac.cn/source/publication/2015/2015-001.pdf
September Calendar of events Would you like to promote events or new publications of your organisation in health highlights? Please send us your contributions! 6 October 2010: Alcolock seminar in the European parliament (Brussels, 6-9 October 2010: 13th European Health Forum (Gastein, Austria)
Job Creation and This Report has been prepared by the Competitiveness Working Group of the European Round Table, inspired by the findings of an ERT Colloquium on "Job Creation through Innovation and Competitiveness" hosted in Brussels on 19 May 1998 by Baron Daniel Janssen on behalf of the ERT. The Colloquium addressed a distinguished audience of senior decision-makers drawn from governments,








