Levitra enthält Vardenafil, das eine kürzere Wirkdauer als Tadalafil hat, dafür aber schnell einsetzt. Männer, die diskret bestellen möchten, suchen häufig nach levitra kaufen ohne rezept. Dabei spielt die rechtliche Lage in der Schweiz eine wichtige Rolle.
Bia.unibz.it

This article was downloaded by:[ABM Utvikling STM / SSH packages]On: 10 April 2008Access Details: [subscription number 787564630]Publisher: Informa HealthcareInforma Ltd Registered in England and Wales Registered Number: 1072954Registered office: Mortimer House, 37-41 Mortimer Street, London W1T 3JH, UK
Microbial Ecology in Health andDisease
Publication details, including instructions for authors and subscription information:
Low prevalence of blaTEM genes in Arctic environments
and agricultural soil and rhizosphere
Lorenzo Brusetti a; Trine Glad b; Sara Borin a; Petter Myren bc; Aurora Rizzi a;
Pål J. Johnsen b; Phil Carter c; Daniele Daffonchio a; Kaare M. Nielsen bd
a Department of Food Science, Technology and Microbiology (DISTAM), Universityof Milan, Milan, Italy
b Department of Pharmacy, Faculty of Medicine, University of Tromsø, Tromsø,Norway
c ESR Kenepuru Science Centre, Porirua, New Zealand
d Norwegian Institute of Gene Ecology, Tromsø, Norway
First Published on: 07 February 2008
To cite this Article: Brusetti, Lorenzo, Glad, Trine, Borin, Sara, Myren, Petter, Rizzi, Aurora, Johnsen, Pål J., Carter,
Phil, Daffonchio, Daniele and Nielsen, Kaare M. (2008) 'Low prevalence of blaTEM genes in Arctic environments and
agricultural soil and rhizosphere', Microbial Ecology in Health and Disease, 20:1, 27 - 36
To link to this article: DOI: 10.1080/08910600701838244
URL:
PLEASE SCROLL DOWN FOR ARTICLE
Full terms and conditions of use:
This article maybe used for research, teaching and private study purposes. Any substantial or systematic reproduction,re-distribution, re-selling, loan or sub-licensing, systematic supply or distribution in any form to anyone is expresslyforbidden.
The publisher does not give any warranty express or implied or make any representation that the contents will becomplete or accurate or up to date. The accuracy of any instructions, formulae and drug doses should beindependently verified with primary sources. The publisher shall not be liable for any loss, actions, claims, proceedings,demand or costs or damages whatsoever or howsoever caused arising directly or indirectly in connection with orarising out of the use of this material.
Microbial Ecology in Health and Disease. 2008; 20: 27�36
Low prevalence of blaTEM genes in Arctic environments andagricultural soil and rhizosphere
LORENZO BRUSETTI1, TRINE GLAD2, SARA BORIN1, PETTER MYREN2,3,AURORA RIZZI1, PA
˚ L J. JOHNSEN2, PHIL CARTER3, DANIELE DAFFONCHIO1 &
KAARE M. NIELSEN2,4
1Department of Food Science, Technology and Microbiology (DISTAM), University of Milan, Milan, Italy, 2Department ofPharmacy, Faculty of Medicine, University of Tromsø, Tromsø, Norway, 3ESR Kenepuru Science Centre, Porirua, NewZealand and 4Norwegian Institute of Gene Ecology, Science Park, Tromsø, Norway
AbstractThe prevalence of blaTEM genes conferring ampicillin resistance (Ampr) in different soils was determined to clarify theenvironmental distribution of resistance determinants of major clinical importance. Samples were collected from 14 sites inNew Zealand, mainland Norway, Svalbard, and 2 soil microcosms made of compost purchased in Italy. The Ampr bacteriarepresented 1.7�100% of the cultivable microflora with an average of 28%. Approximately 1200 Ampr isolates were further
Downloaded By: [ABM Utvikling STM / SSH packages] At: 10:03 10 April 2008
analyzed. Although �50% of the resistant isolates were capable of b-lactam-ring (nitrocefin) degradation, none carried aPCR-detectable blaTEM gene. The proportion of blaTEM genes in the culturable Ampr isolates was B0.07%. The overallblaTEM gene prevalence was determined by blaTEM-specific PCR of DNA extracted directly from the environmental sample.
DNA hybridization was performed on selected samples with a detection limit of �11 blaTEM genes per PCR sample. Ouranalysis indicates that the prevalence of blaTEM carrying bacteria is B1 per 1000 to 100 000 bacteria in the samplesanalyzed. The study suggests that blaTEM genes are rare in soil environments, in contrast to their increasing prevalence insome clinical and commensal bacterial populations. The frequent observation of nitrocefin-degrading capacity among thesampled isolates suggests that other mechanisms conferring enzyme-mediated resistance to b-lactam antibiotics arewidespread in Arctic and agricultural soil environments.
Key words: ampicillin resistance, blaTEM alleles, non-selective environments, natural reservoirs, soil, geographic distribution
fish farms, sewage, drinking water, polluted rivers,and food. The focus of most of these investigations
Bacterial resistance to antimicrobials in previously
has been on the phenotypic characteristics of identi-
susceptible pathogenic species can develop after
fied antibiotic-resistant bacteria (4). With the excep-
spontaneous mutation or horizontal acquisition of
tion of the environmental distribution of some
antimicrobial resistance determinants present inother bacterial populations, species, and environ-
aminoglycoside resistance genes (5�7), the broader
ments (1). Several studies suggest that some resis-
natural prevalence of antibiotic resistance genes is
poorly described in non-clinical environments.
populations of no clinical significance, i.e. the
Among the most clinically and economically
determinants are present in environments not be-
important antibiotic resistance genes are those en-
lieved to encounter significant exposure to, and
coding the b-lactamases (bla genes) (8) producing
selection imposed by, pharmaceutically produced
high level resistance to b-lactam antibiotics, the most
antibiotics (1�3). The description of antibiotic-
widely used antibiotics in clinical and veterinary
resistant bacteria in non-clinical environments has
practice (9,10). A variety of transferable genes
traditionally been limited to those environments
encoding b-lactamase activity have been described
important for human health, such as farm animals,
in clinical environments including blaCTX-M, blaGES,
Correspondence: K.M. Nielsen, Department of Pharmacy, Faculty of Medicine, University of Tromsø, 9037 Tromsø, Norway. Tel: � 47 77646165.
Fax: � 47 77646151. E-mail: [email protected]
(Received 12 February 2007; accepted 2 November 2007)
ISSN 0891-060X print/ISSN 1651-2235 online # 2008 Taylor & FrancisDOI: 10.1080/08910600701838244
L. Brusetti et al.
blaHER, blaOXA, blaOXY, blaSED, blaSHV, blaSPM,
Arctic Svalbard islands (Norway). In addition, two
blaVEB, blaVIM, and ampC alleles. Among the most
soil microcosms kept in a greenhouse with bulk soil
common bla genes is the blaTEM-1 gene, the first
or with maize plants were sampled over time in Italy
described bla gene and a representative of the blaTEM
(Table I). The soil microcosms were made from
group that now consists of almost 150 different
organic compost (Technic no. 7, Potgrondbedrijf
alleles, all encoding different amino acid polymorph-
Vrienzenveen BV, The Netherlands); the composi-
isms that extend their substrate range (http://
tion was described by Brusetti et al. (15). The plant
soil microcosm was prepared by seeding a commer-
variants of the bla
cial maize cultivar (Tundra) in circular-section pots
TEM alleles have only been found
in clinical isolates and are likely emerging as a result
containing organic soil compost and cultivated in a
of point mutations and directional selection. Specific
greenhouse as described previously (15). Bulk soil
ampicillin resistance-encoding bla
microcosms, without plants, were kept under the
TEM alleles are also
present in various bacterial cloning vectors such as
same conditions.
the pUC series, and have been inserted in sometransgenic plant cultivars including commercially
Determination of CFUs of bacteria
approved maize lines (e.g. event Bt176).
Colony-forming units (CFUs) were determined for
TEM alleles, and the TEM-1 allele in
particular, are commonly encountered in clinical
the cultivable aerobic copiotrophic bacteria and for
environments, among pathogenic and commensal
the ampicillin-resistant (Ampr) aerobic copiotrophic
Enterobacteriaceae present in the intestinal tract
bacteria for all environments. Ampr bacteria were
(11,12), in bacterial cloning vectors, and in some
grown on 50 mg/l of ampicillin (Sigma) on agar
transgenic plant varieties (13), few studies have
plates solidified with 15 g/l of BactoAgar (Difco).
determined the non-clinical distribution and poten-
Aerobic copiotrophic bacterial CFUs were deter-
tial natural reservoirs of this important gene, e.g. in
mined on Plate Count Agar medium (Difco) sup-
complex environments such as soil and plant rhizo-
plemented with 100 mg/l of cycloheximide (Fluka),
sphere (14). An expanded study of the ecology of
and enumerated after incubation for 2 days at 308C.
Downloaded By: [ABM Utvikling STM / SSH packages] At: 10:03 10 April 2008
The soil samples from Tromsø, Skibotn, and Sval-
TEM alleles in non-clinical settings will improve our
bard were grown on Plate Count Agar medium
understanding of the origins, environmental dy-
without cycloheximide and enumerated after incu-
namics, and evolution of antibiotic resistance deter-
bation for 3 days at 208C. For the CFUs, means and
minants. In this study, the prevalence of blaTEM
standard deviations (SD) were calculated on the
variants in cultivable and uncultivable ampicillin-
basis of three replicates (five to nine replicates for
resistant bacteria populations was determined from a
Tromsø samples), and analysis of variance of CFU/g
range of geographically diverse soils, composts, and
fresh or dry weight was carried out (Table II).
rhizosphere samples. Material was collected from
Individual means in the ANOVA analysis were
three different agricultural soils and pasture land in
compared using Tukey's test.
New Zealand, six pristine environments in Tromsøand Svalbard, Norway, five farming soils and com-posts made from organic waste from Northern
Identification of ß-lactamase activity with the nitrocefin
Norway, and two soil microcosms made of compost
purchased in Italy. The proportions of ampicillin-
A nitrocefin disk test was used to determine if an
resistant bacteria, the nitrocefin-positive, and the
Ampr isolate had extracellular b-lactamase activity. A
blaTEM-specific PCR-positive isolates were deter-
stock solution (0.5 g/l) of nitrocefin (chromogenic
mined from the cultivable fraction. The potential
cephalosporin compound 87/312, Glaxo) was pre-
presence of unidentified blaTEM-carrying bacteria
pared by adding 9.5 ml of 0.1 M phosphate buffer,
was examined with blaTEM-specific PCR of total
pH 7.0, to 0.5 ml of nitrocefin stock solution (5 mg
DNA extracted from the various samples. We
dissolved in 0.5 ml dimethylsulfoxide). Ten ml of the
provide an estimate of the maximum prevalence of
solution was added to single colonies of each isolate
the blaTEM alleles in the environmental samples
and a color change from yellow to pink within 30
min after application indicated b-lactamase activity.
PCR amplification of potential bla
Materials and methods
TEM genes in the Ampr
Sampling of bulk and rhizosphere soil
Single colonies were used for DNA extraction using
Soils and rhizosphere were sampled from 14 differ-
a boiling lysis method (16). Genomic DNA of the
ent sites in New Zealand, Northern Norway, and the
isolates from the Italian microcosms was extracted
Prevalence of blaTEM genes in soil
Table I. Origin and characteristics of the environmental samples.
Location and description*
Bulk commercial organic compost kept in pots
Maize grown in pots containing commercial organic compost
Ryegrass and white clover grown in untreated soil of volcanic origin
Grass grown in untreated sandy loam soil used for pastoral farming
Browntop, sweet vernal and flatweed grown in untreated agricultural soil
Tromsø; barley, ryegrass, and peas in organic farming agricultural soil
Tromsø; barley, ryegrass, and peas in conventional farming, agricultural soil
Tromsø; barley in organic farming, agricultural soil
Tromsø; barley in conventional farming, agricultural soil
Skibotn; compost made of household waste from Northern Norway
Skibotn; organic soil in low alpine zone
˚ lesund; free soil without vegetation nearby Italian Arctic Research Base
Midtre Love´nbreen; free soil without vegetation in front of glacier moraine
Stuphallet plateau; Saxifraga foliolosa grown in pristine soil
Midtre Love´nbreen; Silene acaulis grown in pristine soil in front of glacier moraine
Glacier sediments
Midtre Love´nbreen; sediments in cryoconite holes
*Italy: samples from compost soils (Technic no. 7, The Netherlands) kept in a greenhouse in Milan were collected from six pots withcultivated maize (rhizosphere samples) and from three pots without cultivated maize (bulk soil samples). For each pot, two samples weretaken after 30 or 100 days of plant growth or incubation. The soil microcosms were sampled as described by Brusetti et al. (15). NewZealand: for the rhizosphere samples, the grass layer was removed and soil from right under the grass and to 10 cm depth was kept. Soilsamples were homogenized by sieving through a 2 mm sieve and stored at 48C (up to 1 week) in double plastic bags before further analysis.
Samples were obtained in October and November 2003 (late spring and early summer in the Southern hemisphere). Norway: the sampleswere collected as follows. For bulk soil, the surface was removed and three replicate samples were collected from 1�5 cm depth into a sterilecontainer. For the rhizosphere, three plants were collected in sterile containers and transported to the laboratory. The soil fraction attachedto the roots was collected in sterile containers by the use of a sterile spatula. Cryoconite holes (water-filled cylindrical melt-holes on glacialice surface) were sampled by removing water and stones and collecting the bottom sediments with a sterile spatula. The Tromsø and Skibotn
Downloaded By: [ABM Utvikling STM / SSH packages] At: 10:03 10 April 2008
samples were analyzed immediately after being returned to the laboratory (within 5 h). The Svalbard samples were stored at 48C untilanalysis (within a few days). The Svalbard sampling was done in August 2004 and the Tromsø and Skibotn samplings in September 2005.
with a CTAB method (17). To confirm the absence
728C for 10 min. For all blaTEM PCR analyses, the
of PCR inhibitory substances in the genomic DNA
primers BlaF and BlaR (Table III) were used to
of the various single isolates, PCR amplification of
amplify a product of 828 bp (TEM-1 allele). The
16S rRNA gene was performed on a subset of the
specificity of the primers was confirmed by ‘in silico'
DNA extracts with the primers 16S-27F and 16S-
1494R (Table III). The genomic DNA extracted
infx) and by aligning the primer binding region of
through the CTAB method was confirmed as
approximately 100 sequence polymorphic TEM-
suitable for PCR analysis by performing PCR of
alleles (Table IV).
the bacterial 16S-23S rRNA gene intergenic tran-
The following controls were used: five strains of
scribed spacers (ITS-PCR) with primers ITSF and
Escherichia coli carrying the bla alleles TEM-1,
ITSReub (18), as described by Daffonchio et al.
TEM-3, TEM-6, TEM-9, and TEM-10 as positive
controls, and one strain carrying the SHV-2 allele as
The amplification of blaTEM alleles in individual
negative control. The blaSHV-2 gene shares about
bacterial isolates was performed in a reaction mix-
70% of nucleotide similarity with the blaTEM gene
ture containing 1 �HotStartTaq DNA master mix,
cluster. All strains were kindly provided by A.
0.2 mM of each primer, and 2 ml of the crude DNA
Sundsfjord, University Hospital of North-Norway,
solution in a final volume of 30 ml. Bacterial isolates
Tromsø, Norway. Two strains, Pseudomonas putida
from the microcosm samples were in addition
ET-B12 and Flavobacterium sp. ET-N11, found to be
analyzed by PCR using a different reaction mixture
resistant to ampicillin but without an identifiable
containing 1 � PCR buffer (Pharmacia), 1.5 mM
blaTEM gene, and previously isolated from a soil
MgCl2, 0.10 mM dNTPs, 0.2 mM of each primer, 1
microcosm (Italy), were used as positive controls
U of native Taq polymerase (Pharmacia), and 2 ml of
after they had been transformed by electroporation
genomic DNA in a final volume of 50 ml. Reactions
with plasmid pZR80 carrying the blaTEM-1 gene
were denatured at 958C for 15 min and then
conferring resistance to ampicillin (17,20). To con-
subjected to 30 cycles of 948C for 45 s, 618C for
firm the presence of the pZR80 plasmid in the two
45 s, and 728C for 1 min, with a final extension at
electroporated strains, direct sequencing of the
L. Brusetti et al.
Table II. Colony forming units (log10) of total and ampicillin-resistant bacteria (50 mg/l) obtained from the various environments.
Ampr copiotrophic
Italian soil microcosm
Bulk soil, day 30
Bulk soil, day 100
Maize rhizosphere, day 30
Maize rhizosphere, day 100
New Zealand agricultural soils
Horotiu, rhizosphere
Otaki, rhizosphere
Templeton, rhizosphere
Tromsø, bulk soil, organic farming
Tromsø, bulk soil, conventional farming
Tromsø, rhizosphere, organic
Tromsø, rhizosphere, conventional
Skibotn, low alpine soil
Svalbard soils and sediments
˚ lesund, bulk soil
Lovenbreen, bulk soil
Stuphallet, rhizosphere
Lovenbreen, rhizosphere
Lovenbreen, glacier sediments
*Total copiotrophic bacteria and Ampr bacteria measured as log10 CFU/g (dry weight) of soil. For the Svalbard samples, the CFU were
Downloaded By: [ABM Utvikling STM / SSH packages] At: 10:03 10 April 2008
calculated as log10 CFU/g (fresh weight) of soil. Mean values are given with standard deviation (SD) based on three replicates; five to ninereplicates for Tromsø samples.
$Percentage of Ampr CFU of the total culturable bacterial population.
flanking regions of the blaTEM gene were performed
were deposited in the EMBL nucleotide sequence
using the sequencing primers TemI3, TemI5a, or
database (GenBank/EMBL/DDBJ) under the acces-
TemI5b (Table III).
sion numbers AM261986 to AM262113.
Sequencing of the 16S rRNA gene of Ampr isolates
Extraction and analysis of DNA from soil
To describe some of the diversity of Ampr bacterial
Total DNA from 11 different sample sites was
phenotypes present in the Italian soil microcosm,
extracted from 0.5 g of material using the Bio101
288 isolates were analyzed with ITS-PCR (see
FastDNA Spin Kit for soil (Q-BioGene) according
above) (21). One isolate of each of the ITS-PCR
to the manufacturer's instructions. Further purifica-
haplotypes was further identified by partial 16S
tion and concentration of eluted DNA was per-
rRNA gene sequence analysis described by Brusetti
formed with the QIAamp DNA Stool Mini Kit
et al. (22) using primer 16S-926F (Table III). The
(Qiagen). Soil DNA was finally eluted in 30 ml of
nucleotide sequences of the partial 16S rRNA genes
TE buffer, pH 8. The general suitability of DNA for
Table III. PCR and sequencing primers used in this study.
Primer sequence (5?-3?)
Prevalence of blaTEM genes in soil
Table IV. Identified nucleotide polymorphisms between the BlaF and BlaR primers used and some published blaTEM-alleles*.
BlaF primer (5?-3?)
blaTEM-1b, 1d, 1e, 1g, 1h, 5, 10, 15, 18-19, 27-28, 33, 35, 47, 50, 55, 61, 68, 70, 76-77, 79, 88,
92, 95, 102, 104-107
BlaR primer (5?-3?)
*The primer sequences are identical to the blaTEM-1a allele. In addition to the alleles blaTEM-1a to -1h and blaTEM-116, the alignment includedall alleles up to blaTEM-107, except blaTEM-14, -23, -37, -58, -62, -64, -67, -69, -75, -100, -103. Only alleles with nucleotide polymorphisms in theprimer binding regions are shown.
PCR was confirmed with amplification of the 16S
pUC18 molecules. The variation in the pUC18 copy
rRNA gene. The PCR mixture contained 1 �Hot-
number, as affected by variation in individual DNA
StartTaq master mix, 0.3 mM of each primer, 1 �Q-
sample measurements, ranged from 1.4 �1011 to
solution (Qiagen), 0.1 mg/ml bovine serum albumin,
3.4 �1011 for the estimated 1.7 �1011 copies, and
5% dimethyl sulfoxide, 2 mM of MgCl2, and 2 ml of
from 2 to 5 copies for the estimated 3 copies.
extracted DNA (100 ng/ml) in a final volume of
Stochasticity in the pUC18 distribution during
30 ml. The number of cycles was increased to 33.
dilutions makes the lower estimate less precise.
The PCR targeting of blaTEM genes from total DNA
The detection limit of the blaTEM-specific PCR was
was performed with the primers described in Table
also determined without soil DNA present using a
III and 2 ng of DNA per reaction.
range of pUC18 dilutions. The detection limit wasdetermined in total DNA extractions of the soilsamples from Tromsø, Skibotn, and Svalbard.
Downloaded By: [ABM Utvikling STM / SSH packages] At: 10:03 10 April 2008
Southern blotting of blaTEM PCR products
Agarose gels of blaTEM-specific PCRs performed onsome of the DNA samples were blotted overnight
onto positively charged nylon membranes (Pharma-
Bacterial counts of the culturable bacterial fraction
cia) by capillary transfer (17). The membrane wastreated with UV light for 4 min and hybridized
The bacterial counts (CFUs) of the total and the
overnight at 428C in 50% (v/v) formamide with the
Ampr fraction of aerobic copiotrophic bacteria were
corresponding DIG-labelled blaTEM-1 variant. Two
determined in all samples (Table II). The cultivable
stringent washes were performed at 658C for 15 min.
copiotrophic bacteria ranged between 4.7 and 8.6
Prehybridization, hybridization, washes, and chemi-
log10 CFU/g dry weight for Italian compost and
luminescent detection were performed using the
mainland Norway samples New Zealand soil samples,
DIG DNA labelling and detection kit following the
and between 4.3 and 6.1 log10 CFU/g fresh weight for
supplier's instructions (Boehringer Mannheim). The
Svalbard. The proportion of ampicillin-resistant bac-
probe was DIG-labeled by PCR using the conditions
teria (measured as growth on 50 mg/l ampicillin)
described for the blaTEM PCR.
ranged from 1.7% to 100% of the culturable bacterialfraction (Table II). The New Zealand soil sampleswere characterized by a relatively high percentage of
Limit of detection for blaTEM genes in soil DNA
Ampr bacteria (between 31.9% and 54.9%), whereas
To determine the detection limit of the blaTEM
the soils from Northern Norway (Tromsø and Ski-
amplification of total DNA, 2 ng of total soil DNA
botn) had lower levels of resistant bacteria (8.8�
was added to each reaction spiked with decreasing
16.1%). The observable proportion of Ampr cultur-
amounts of commercially available pUC18 plasmids
able bacteria in the samples from Svalbard ranged
(Invitrogen) from 1.7 �1011 to 3 molecules. To
from 20.2% in bulk soil to 100% in glacier sediments.
describe the variation in the calculated number of
Compost samples such as the organic plant compost
pUC18 molecules, the DNA concentration of un-
from Skibotn, Norway, and the Italian bulk soil
diluted and 10-fold dilution of pUC18 were mea-
microcosm had the lowest percentage of Ampr
bacteria (1.7% and 3.5%, respectively). In the bulk
(NanoDrop Technologies). The highest and lowest
soil microcosm (Italy), the relative percentage of
values of 18 samples taken from the DNA stock were
Ampr bacteria increased significantly from 3.5% to
used to calculate the outer concentration range of
9.4% (p �0.00001), over the 100 days incubation
L. Brusetti et al.
period. In the plant soil microcosms, the percentage
PCR targeting of blaTEM genes in the Ampr isolates
of Ampr bacteria decreased significantly from 17.2%
The suitability for PCR analysis of the DNA
to 11.6% over the 100 days incubation period (p �
prepared from the 1329 Ampr bacterial isolates was
0.00004). Despite the relative percentage changes,
confirmed with PCR amplification of the 16S rRNA
the overall levels of Ampr bacteria changed little, from
gene or by ITS-PCR. Of the 1329 DNA prepara-
5.2�5.6 log10 CFU/g dry weight to 5.0 log10 CFU
tions tested, 1198 were positive, indicating that
over the time course. Significant differences (p B
bacterial DNA was amplifiable in 90.1% of the
0.05) between plate counts with or without ampicillin
samples. Subsequently, 1198 isolates were screened
were found in all the samples except for two soil and
for the presence of bla
sediment samples from Svalbard, both with an Ampr
genes with primers
designed for the TEM-1 allele and derivatives.
proportion �60%.
PCR of the different TEM-harboring E. coli strainsand of the two pZR80-transformed strains was
Phenotypic analysis of the Ampr bacterial isolates
positive, while amplification of the SHV-2 genegave a faint band (data not shown). Of the 1198
A total of 1329 Ampr bacterial isolates was obtained
environmental isolates analyzed by PCR, none
from the various environments. The nitrocefin test
produced visible bla
was performed on 1185 isolates (Table V). The
TEM amplicons (Table V). In
silico PCR, absence of primer mismatches to the
cephalosporin nitrocefin was cleaved by 97.7% of
most relevant TEM alleles (Table IV), and amplifi-
the bacterial isolates, excluding the New Zealand
cation of the positive controls demonstrate the broad
soils and the Svalbard glacier sediment samples. For
ability of the primers used to detect relevant TEM
the New Zealand soil samples, the nitrocefin-de-
alleles across phylogenetically diverse groups, if
grading Ampr bacteria represented only a minor
present in the isolates.
fraction, between 18.6% and 30.7% of the totalAmpr isolates. No nitrocefin-degrading bacteria wereidentified among the isolates in Svalbard cryoconite
Identification of Ampr isolates through partial 16S
holes containing glacier sediments, despite the ob-
Downloaded By: [ABM Utvikling STM / SSH packages] At: 10:03 10 April 2008
served 100% proportion of Ampr cultivable bacteria.
To provide some insight into the species diversity of
Table V. Nitrocefin substrate test and blaTEM targeted PCR
the Ampr bacteria isolated in a specific soil environ-
analysis of single Ampr bacterial isolates from each environment.
ment, a total of 288 Ampr isolates from the soilmicrocosms made of purchased compost (Italy) were
Isolates positive/tested
grouped into 139 haplotypes by ITS-PCR finger-printing analysis (19). The 16S rRNA gene se-
quences subsequently obtained covered between
Italian soil microcosm
240 and 570 bp, with an average of 506 bp.
Bulk soil, day 30
At day 30 of the greenhouse incubation, the Ampr
Bulk soil, day 100
bulk soil isolates were characterized by the presence
Maize rhizosphere, day 30
of 13 different genera. Of the isolates examined,
Maize rhizosphere, day 100
44% were b-Proteobacteria, mostly belonging to the
New Zealand agricultural soils
genera Acidovorax and Pandoraea; 21% of the isolates
Horotiu, rhizosphere
Otaki, rhizosphere
were affiliated to a-Proteobacteria of the genera
Templeton, rhizosphere
Ochrobactrum, Caulobacter, and Zooglea. g-Proteo-
Norwegian mainland soils
bacteria was represented by 15% of the isolates. One
Tromsø, bulk soil, organic farming
isolate of the Actinobacteria (Streptomyces sp.) was
Tromsø, bulk soil, conventional
found. At day 100, the majority of the isolates
belonged to 4 different species of Pseudomonas (P.
Tromsø, rhizosphere, organic
alcaligenes, P. jessenii, P. putida, and P. resinovorans),
Tromsø, rhizosphere, conventional
although 25 different Pseudomonas species were
Skibotn, low alpine soil
identified among the isolates. The other sequenced
Svalbard soils and sediments
isolates were grouped to Acidovorax spp., Chryseo-
˚ lesund, bulk soil
bacterium sp., Flavobacterium sp., Mesorhizobium sp.,
Lovenbreen, bulk soil
Roseateles sp., and Stenotrophomonas sp.
Stuphallet, rhizosphere
At day 30 in the maize rhizosphere microcosm
Lovenbreen, rhizosphere
samples, the bacterial isolates sequenced mainly
Lovenbreen, glacier sediments
belonged to b- and g-Proteobacteria and to the
Bacteroidetes/Chlorobi group. The Bacteroidetes/
ND, not determined.
Chlorobi group bacteria were mostly represented
Prevalence of blaTEM genes in soil
by Dyadobacter fermentens and Flexibacter sanctispecies (10% of the strains). Other genera repre-sented were Acidovorax, Flavobacterium, Pandoraea,Pseudomonas, and Stenotrophomonas. In the rhizo-sphere of maize sampled at day 100, 56% of the 16Ssequenced bacteria were affiliated to the g-Proteo-bacteria class, exclusively to the genera Pseudomonasand Stenotrophomonas. The species P. fluorescens, P.
jessenii, P. monteilii, P. pseudoalcaligenes, P. putida, andP. resinovorans were identified. Other bacterial generawere represented by Azospirillum sp., Chryseobacter-ium sp., Flavobacterium sp., Mesorhizobium sp.,Ochrobactrum sp., and Variovorax sp. Some of theabove species or genera are known to be b-lactamasecarriers (23,24).
Detection limit of the PCR targeting of blaTEM genes
Eleven soil, rhizosphere, and sediment samples fromNorway were screened for the presence of blaTEMgenes in extracted total DNA. No visible amplifica-tion products of the blaTEM genes were obtainedusing 2 ng of extracted DNA in the reactions. DNA
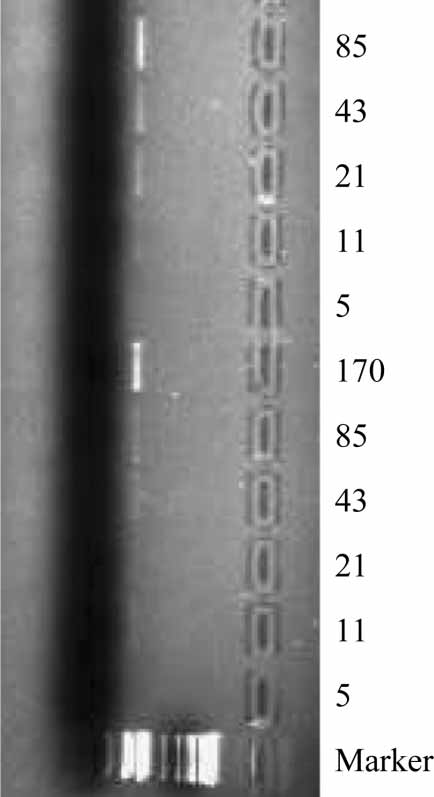
Figure 1. PCR with blaTEM-specific primers of dilution series of
sequence alignment, and the PCR-positive controls
pUC18 DNA (estimated copy number of 170 to 5), in the
(TEM-1, -3, -6, -9, -10), as well as spiked soil
presence of soil DNA extracts. Lanes 1�5, with soil DNA from
samples (pUC18) demonstrated the broad ability of
organic farming in Tromsø; lanes 6�10, DNA from compost from
Downloaded By: [ABM Utvikling STM / SSH packages] At: 10:03 10 April 2008
Northern Norway; 12, DNA fragment size ladder.
the TEM-specific primers used to detect relevantTEM alleles in the DNA samples analyzed. In 10 of
genes for experimental field releases of genetically
11 samples, the detection limit of the blaTEM
modified organisms (14). However, few experimen-
amplification products visualized in agarose gels
tal data exist on the environmental distribution of
was 21 (range 17�43, see Materials and methods)
pUC18 molecules (Figure 1). The extracted DNA
TEM genes in natural environments, such as in
food, feed, water, bulk soil, rhizosphere, compost,
from low alpine soil in Skibotn, Norway, did not
and pristine environments (14).
yield a satisfactory detection limit when spiked with
In this study, the prevalence of bla
the pUC18 plasmid. To further increase the sensi-
was surveyed in microbial populations of a variety of
tivity, Southern hybridizations were done on thePCR solutions of some of the soil DNA samples
environments. The concentrations of cultivable
(Figure 2). Very faint bands were observed in the
Ampr bacteria in all samples was high, typically
blotted PCR solution from Ny A
˚ lesund and Tromsø
103�108 CFU per dry weight gram of sample; in
bulk soil, rhizosphere from Lovenbreen, and from
some samples they represented �50% of the total
conventional farming land from Tromsø. Southern
culturable microbial population (Table II). The high
blotting of PCR products from solutions with
proportion of Ampr bacteria already present in the
decreasing amounts of pUC18 target DNA yielded
samples suggests that in situ or in vitro enrichment
a clear signal with 85 molecules and was positive
(with ampicillin selection) would enhance our cap-
with as low as 11 pUC18 copies (Figure 2).
ability to detect blaTEM-positive bacteria only ap-proximately twofold, assuming uniform communityresponse to the enrichment conditions.
Although the majority of �1100 isolates tested
were nitrocefin-degrading (hence capable of cleav-
TEM types are among the earliest described b-
lactamase encoding genes (11; http://www.lahey.org/
ing the b-lactam ring), bacterial isolates carrying
Studies/), they are frequently used in cloning vectors
blaTEM-alleles could not be identified in the analyzed
in molecular biology laboratories (13), and are
environmental isolates (Table V). Lower proportions
present in some of the first generation genetically
of nitrocefin-degrading bacteria were seen in New
modified plants (14,25). The broad environmental
Zealand soil samples and in the sediment samples
distribution of specific blaTEM alleles has been used
from cryoconite holes in glaciers on Svalbard. In the
as an argument for their continued use as marker
latter the entire cultivable microbial population was
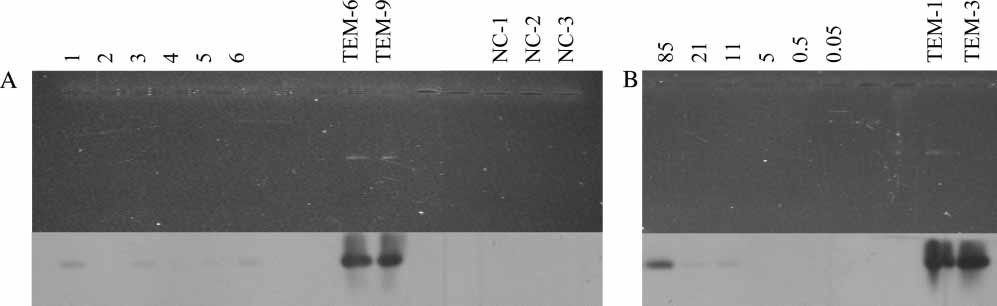
L. Brusetti et al.
Figure 2. Southern blotting of agarose gel separated PCR amplified DNA with blaTEM-specific primers. (A) Lane 1, Ny A
˚ lesund, bulk soil
(Svalbard); 2, Stuphallet, rhizosphere (Svalbard); 3, Lovenbreen, rhizosphere (Svalbard); 4, Tromsø, bulk soil, organic farming; 5, Tromsø,bulk soil, conventional farming; 6, Tromsø, rhizosphere, conventional farming. (B) PCR of dilution series of pUC18 DNA from anestimated copy number of 85 to 0.05. Lanes TEM-1, TEM-3, TEM-6, and TEM-9 are positive controls with 3 ng of PCR products ofdifferent variants of blaTEM genes, and NC lanes are negative controls (no DNA added to the PCR reaction).
Ampr, yet nitrocefin-degrading bacteria were not
despite not providing evidence for direct transfer of
found (Table V). The high proportion of bacteria
resistance elements � they proposed a previously
with extracellular b-lactamase activity in environ-
under-appreciated density and concentration of en-
ments without known or expected prior exposure to
vironmental antibiotic resistance determinants.
pharmaceutically produced b-lactam compounds
Our data from soil microcosms (made from
suggests that antibiotic-degrading capacities are
commercially available compost) incubated under
also important in microbial communities unaffected
controlled greenhouse conditions for 3 months
by humans. The molecular and ecological basis for
indicate that the relative levels of Ampr bacteria
this observation needs to be determined and the
change over time. After 1 month, the relative
natural substrates for these enzymes need to be
percentage of Ampr bacteria in the bulk soil compost
identified to advance our understanding and pre-
was about four times lower than in the same soil 3
dictive ability of the capacities of broadly distributed,
months later. An opposite and also significant
Downloaded By: [ABM Utvikling STM / SSH packages] At: 10:03 10 April 2008
naturally occurring enzymes to act on pharmaceuti-
difference was observed for the proportion of Ampr
bacteria in maize plant-containing soil microcosms
sequence-divergent representatives of the chromoso-
(see Table II). Temporal fluctuations in the resis-
mally localized ampC gene to be responsible for
tance proportions were also described for tetracy-
some of the resistant phenotypes observed.
The fraction of Ampr bacteria reported here is
bacteria (7,30). The observed fluctuations in resis-
lower than the percentage of Ampr bacteria found in
tance levels over time probably reflect population
other soil and rhizosphere samples (26,27). A recent
dynamic processes among bacterial species as a
study by Badosa et al. (28) indicated that the Ampr
result of competition without necessarily reflecting
bacterial fraction in some maize fields in Spain
directional selection of resistance traits.
represented between 77% and 88% of the total
The BLAST analysis of 16S rRNA genes amplified
cultivable bacterial population in the rhizosphere,
from 139 Ampr haplotypes revealed a wide variety of
and between 68% and 88% in bulk agricultural soil.
ampicillin-resistant species (MIC �50 mg ampicil-
These latter high proportions are close to those
lin/l), belonging to a-, b-, and g-Proteobacteria,
observed for the rhizosphere samples from Svalbard.
Sphingobacteria, the Bacteroidetes/Chlorobi group,
Several other studies of the distribution of pheno-
and to Actinobacteria. Although the TEM-1 allele
typic antibiotic resistance patterns have been re-
has mainly been linked to commensal and clinical
pathogenic isolates identified as Capnocytophaga
bacteria in Danish farmlands soils was reported to
ochracea, Enterobacter cloacae, E. coli, Klebsiella oxy-
be between 0.1% and 7.1%, with an average of 2.1%
toca, Klebsiella pneumoniae, Morganella morganii,
(30). Similar percentage levels were found for
Proteus mirabilis, Providencia stuartii, Pseudomonas
streptomycin-resistant bacteria in bulk and rhizo-
aeruginosa, Zymomonas mobilis and others, non-
sphere soils (7). However, higher proportions of
TEM-like b-lactamase-producing genes have been
gentamicin-resistant (exact numbers not given) and
described in some soil bacteria. The class B b-
kanamycin-resistant (between 0.01% and 38.6%)
lactamases have been found in B. cereus (31),
bacteria among the cultivable copiotrophic or oligo-
Chryseobacterium meningosepticum (23), and S. mal-
trophic bacteria were found in different European
tophilia (24).
bulk and rhizosphere soils (5,6). D'Costa et al. (29)
The absence of the blaTEM gene in �1100
found high levels of multi-drug-resistant actinomy-
cultured Ampr environmental isolates indicates that
cetes in urban, agricultural, and forest soil and �
the blaTEM genes are rare in soil environments
Prevalence of blaTEM genes in soil
despite the reports of high prevalence of blaTEM-1
tion and not convergent evolution. The few single
genes in some pathogenic bacterial species (11), and
nucleotide polymorphisms (SNPs) present in the
the occurrence of the gene in the digestive system of
TEM alleles strongly suggest a shared recent origin
healthy humans (12). Since no blaTEM-carrying
and subsequent vertical and horizontal dissemina-
bacteria were identified among the examined Ampr
tion among bacterial species. Sequence analysis of a
isolates, it is important to be aware of the limit of
broader set of bacterial species (from various envir-
detection provided by the experimental methods. It
onments) carrying TEM-like alleles is necessary to
can be inferred that the concentration of culturable
resolve the evolution and identity of the original
Ampr bacteria is B0.3% in the Italian soil micro-
bacterial blaTEM-1 gene donor.
B0.2% in the combined New Zealand
agricultural soils, B0.4% in the combined Norwe-
gian soil samples, and B0.3% in the Svalbard
We thank M. Pajoro, A. Pagliuca, and P. Francia for
samples, and not detectable among cultured isolates.
technical assistance in plant cultivation, bacterial
The proportion of blaTEM genes in the combined
counts, and strain isolation performed at the Uni-
samples of all Ampr isolates is B0.07%. The
versity of Milan. This work was supported by the
phenotypic analysis represents 0.001�32.5% of the
Italian Ministry for University and Scientific Re-
amount of culturable Ampr bacteria present in 1 g of
search project ‘Risposta della comunita microbica
sample material. This study is thus comparable to
del suolo a differenti pressioni antropiche: effetti su
the sample sizes of cultured bacteria analyzed in
struttura, dinamica e diversita della microflora' and
previous studies (28,32).
the Fondazione Diritti Genetici, Italy, project ‘Or-
A detection limit of B21 pUC18 copies per PCR
ganismi geneticamente modificati ed alimentazione:
sample (established through spiking of the soil DNA
valutazione degli effetti diretti sull'ospite e sulla
with decreasing concentrations of pUC18) was
microflora intestinale' funded by the Cariplo Foun-
obtained for the total DNA extracted from soil.
dation, Italy. Partial support also came from the EU
Our PCR protocol included 2 ng of DNA from the
project TRANSBAC QLK3-CT-2001-02242. T.G
soil samples. Assuming that all DNA extracted is of
Downloaded By: [ABM Utvikling STM / SSH packages] At: 10:03 10 April 2008
and K.M.N acknowledge financial support from the
bacterial origin, it represents the DNA in maximum
Research Council of Norway. Support for the New
100 000 bacterial cells (32) or in approximately
Zealand work was provided by the Foundation for
0.02�0.2 mg soil (assuming 10�100 mg DNA per g
Research, Science and Technology.
soil). Moreover, since the blaTEM gene is likely to belocalized on plasmids (and assuming 11 or morecopies per bacterial cell) it can be inferred that the
blaTEM gene is present in the samples analyzed atproportions lower than 1 bla
1. Nwosu VC. Antibiotic resistance with particular reference to
TEM-carrying bacterium
soil microorganisms. Res Microbiol. 2001;152:421�30.
per 100 000 bacteria analyzed. However, the detec-
2. Mazel D, Davies J. Antibiotic resistance in microbes. Cell Mol
tion limit in our experimental approach is probably
Life Sci. 1999;56:742�54.
100-fold higher, due to the presence of non-bacterial
3. Seveno N, Smalla K, Van Elsas JD, Collard J-C, Karagouni A,
DNA (e.g. of fungal origin) in the DNA extraction
Kallifidas D, et al. Occurrence and reservoirs of antibiotic
from soil. Thus, our investigation indicates that the
resistance genes in the environment. Rev Med Microbiol.
2002;13:1�13.
prevalence of bla
TEM-carrying bacteria in the sam-
4. Henschke RB, Schmidt FRJ. Screening of soil bacteria for
ples analyzed is B1 per 100 000 to 1 per 1000
plasmids carrying antibiotic resistance. Biol Fertil Soils. 1990;/
This investigation suggests that bla
5. Smalla K, Van Overbeek LS, Pukall R, Van Elsas JD.
Prevalence of nptII and Tn5 in kanamycin-resistant bacteria
rare in the non-clinical environments sampled in our
from different environments. FEMS Microbiol Ecol. 1993;13:
investigations. b-Lactamases such as Tem-1 are
hypothesized to have evolved from the penicillin-
6. Heuer H, Kro¨gerrecklenfort E, Wellington EMH, Egan S,
binding proteins (PBPs) due to directional selection
Van Elsas JD, Van Overbeek L, et al. Gentamicin resistance
on the PBP's active site (changing activity from
genes in environmental bacteria: prevalence and transfer.
FEMS Microbiol Ecol. 2002;42:289�302.
binding and inhibition by penicillin to penicillinase
7. Van Overbeek LS, Wellington EMH, Egan S, Smalla K,
activity and detachment of the enzyme from the
Heuer H, Collard J-M, et al. Prevalence of streptomycin
cytoplasmic membrane) (33,34). The blaTem-1 allele
resistance genes in bacterial populations in European habitats.
was first described in 1965 and is now widespread in
FEMS Microbiol Ecol. 2002;42:277�88.
clinical environments and among Enterobacteria-
8. Bush K. Characterization of b-lactamases. Antimicrob Agents
ceae (35,36). The increased prevalence of the
9. Singh GS. b-Lactams in the new millennium. Part-I: mono-
TEM-1 allele in human-associated bacterial popula-
bactams and carbapenems. Mini Rev Med Chem. 2004;4:69�
tions is due to directional selection and dissemina-
L. Brusetti et al.
10. Kumar K, Gupta SC, Chander Y, Singh AK. Antibiotic use in
23. Rossolini GM, Franceschini N, Riccio ML, Mercuri PS,
agriculture and its impact on the terrestrial environment. Adv
Perilli M, Galleni M, et al. Characterization and sequence of
the Chryseobacterium (Flavobacterium) meningosepticum carba-
11. Livermore DL. b-lactamases in laboratory and clinical
penemase: a new molecular class b-lactamase showing a
resistance. Clin Microbiol Rev. 1995;8:557�84.
broad substrate profile. Biochem J. 1998;332:145
˜ as L, Zarazaga M, Sa´enz Y, Ruiz-Larrea F, Torres C. b-
24. Ullah JH, Walsh TR, Taylor IA, Emery DC, Verma CS,
Lactamases in ampicillin-resistant Escherichia coli isolates
Gamblin SJ, et al. The crystal structure of the L1 metallo-
from foods, humans, and healthy animals. Antimicrob Agents
beta-lactamase from Stenotrophomonas maltophilia at 17 A
resolution. J Mol Biol. 1998;284:125�36.
13. Bensasson D, Boore JL, Nielsen KM. Genes without fron-
25. EFSA. Opinion of the scientific panel on genetically modified
tiers? Heredity. 2004;92:483�9.
organisms on the use of antibiotic resistance genes as marker
14. Nielsen KM, Berdal KG, Kruse H, Sundsfjord A, Mikalsen
genes in genetically modified plants. EFSA J 2004;48:1�18.
A, Yazdankhah S, et al. An assessment of potential long-term
26. Van Dijk P, Van de Voorde H. Sensitivity of environmental
health effects caused by antibiotic resistance marker genes in
microorganisms to antimicrobial agents. Appl Environ Micro-
genetically modified organisms based on antibiotic usage and
resistance patterns in Norway. Oslo, Norway: Norwegian
27. Brønstad K, Drønen K, Øvrea˚s L, Torsvik V. Phenotypic
Scientific Committee for Food Safety Report; 2005. p. 1�62.
diversity and antibiotic resistance in soil microbial commu-
15. Brusetti L, Francia P, Bertolini C, Pagliuca A, Borin S, Sorlini
C, et al. Bacterial communities associated with the rhizo-
nities. J Indust Microbiol. 1996;17:253�9.
sphere of transgenic Bt 176 maize (Zea mays) and its non
28. Badosa E, Moreno C, Montesinos E. Lack of detection of
transgenic counterpart. Plant Soil. 2004;266:11�21.
ampicillin resistance gene transfer from Bt176 transgenic corn
16. Glad T, Klingenberg C, Flægstad T, Ericson JU, Olsvik Ø.
to culturable bacteria under field conditions. FEMS Micro-
Rapid detection of the methicillin-resistance gene, mecA, in
biol Ecol. 2004;48:169�78.
coagulase-negative Staphylococci. Scand J Infect Dis. 2001;
29. D'Costa VM, McGrann KM, Hughes DW, Wright GD.
Sampling the antibiotic resistome. Science. 2004;311:374�7.
17. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman
30. Sengeløv G, Agersø Y, Halling-Sørensen B, Baloda SB,
JG, Smith JA, et al. Current protocols of molecular biology.
Andersen JS, Jensen LB. Bacterial antibiotic resistance levels
New York: John Wiley and Sons; 1994.
in Danish farmland as a result of treatment with pig manure
18. Cardinale M, Brusetti L, Quatrini P, Borin S, Puglia AM,
slurry. Environ Int. 2003;28:587�95.
Rizzi A, et al. Comparison of different primer sets for the
31. Sabath LD, Abraham EP. Zinc as a cofactor for cephalospor-
Automated Ribosomal Intergenic Spacer Analysis (ARISA) of
inase from Bacillus cereus 569. Biochem J. 1966;98:11�3.
Downloaded By: [ABM Utvikling STM / SSH packages] At: 10:03 10 April 2008
complex bacterial communities. Appl Environ Microbiol.
32. Nielsen KM, Townsend JP. Monitoring and modeling hor-
izontal gene transfer. Nature Biotechnol. 2004;22:1110�4.
19. Daffonchio D, Borin S, Frova G, Manachini PL, Sorlini C.
33. Kirby R. Evolutionary origin of the class A and class C beta-
PCR fingerprinting of whole genomes, the spacers between
lactamases. J Mol Biol. 1992;34:345�50.
the 16S and 23S rRNA genes and of intergenic tRNA gene
34. Hall B, Barlow M. Evolution of the serine beta-lactamases:
regions reveals a different intraspecific genomic variability of
past, present and future. Drug Res Updates. 2004;7:111�23.
Bacillus cereus and Bacillus licheniformis. Int J Syst Bacteriol.
35. Amyes SGB. Genes and spectrum: the theoretical limits.
Clinic Infect Dis. 1998;27(Suppl 1):S21�8.
20. Kok RG, Young DM, Ornston LN. Phenotypic expression of
36. Kotra LP, Samama J-P, Mobashery S. b-Lactamases and
PCR-generated random mutations in a Pseudomonas putida
resistance to b-lactam antibiotics. In: Lewis K, Salyers AA,
gene after its introduction into an Acinetobacter chromosome
Taber HW, Wax RG. editors. Bacterial resistance to anti-
by natural transformation. Appl Environ Microbiol. 1999;65:
microbials. New York: Marcel Dekker; 2002. p. 126�9.
37. Ehlers B, Strauch E, Goltz M, Kubsch D, Wagner H,
¨ rtler V, Stanisich VA. New approaches to typing and
Maidhof H, et al. Nachweis gentechnischer vera¨nderungen
identification of bacteria using the 16S�23S rDNA spacer
in mais mittels PCR. Bundesgesundhbl. 1997;4:118�21.
region. Microbiology. 1996;142:3�16.
38. Lane DJ. 16S/23S rRNA sequencing. Nucleic acid techniques
22. Brusetti L, Borin S, Mora D, Rizzi A, Raddadi N, Sorlini C,
in bacterial systematics. In: Stackebrandt E, Goodfellow M.
et al. Usefulness of length heterogeneity-PCR for monitoring
editors. Modern microbiological methods. Chichester, UK: J
lactic acid bacteria succession during maize ensiling. FEMSMicrobiol Ecol. 2006;56:154�64.
Wiley & Sons; 1991. p. 133.
Source: https://bia.unibz.it/bitstream/handle/10863/215/2008-BrusettiMEHDBla-TEM.pdf?sequence=2
Comprehensive Capacity Assessment of Health Laboratory Services in Nepal National Public Health laboratory and WHO-Nepal Page 1 of 62 Comprehensive Capacity Assessment of Health Laboratory Services in Nepal Dr. Palpasa Kansakar, Ph.D. (Microbiology) Mr. Binod Kumar Yadav, M.Sc (Biochemistry) Mr Krishna Rijal, CMLT Page 2 of 62
FRAnCiSley ÁvilA SouzA1, AnA PAulA FARnezi BASSi1, AleSSAndRA MARCondeS ARAnegA1, dAnielA Ponzoni1, gABRielA BuFulin leonARdi2, FeRnAndA BRASil dAuRA JoRge BooS3, eloÁ RodRigueS luvizuTo4, HeloíSA HelenA níMiA5, idelMo RAngel gARCiA JúnioR1 1DDs, Ms, PhD. Professors of the surgery and Integrated Clinic Department at the Araçatuba of Dental school – Univ est Paulista Júlio de



