Levitra enthält Vardenafil, das eine kürzere Wirkdauer als Tadalafil hat, dafür aber schnell einsetzt. Männer, die diskret bestellen möchten, suchen häufig nach levitra kaufen ohne rezept. Dabei spielt die rechtliche Lage in der Schweiz eine wichtige Rolle.
Genome89045 1184.1194
Lack of bystander activation shows that localization exterior to
chromosome territories is not sufficient to up-regulate gene
expression
Céline Morey, Clémence Kress and Wendy A. Bickmore
2009 19: 1184-1194 originally published online April 23, 2009
Genome Res.
This article cites 34 articles, 19 of which can be accessed free at:
Receive free email alerts when new articles cite this article - sign up in the box at the
top right corner of the article or
Genome Research
Copyright 2009 by Cold Spring Harbor Laboratory Press
Lack of bystander activation shows that localizationexterior to chromosome territories is not sufficientto up-regulate gene expression
Ce´line Morey,1 Cle´mence Kress,2 and Wendy A. Bickmore3MRC Human Genetics Unit, Edinburgh EH4 2XU, Scotland, United Kingdom
Position within chromosome territories and localization at transcription factories are two facets of nuclear organizationthat have been associated with active gene expression. However, there is still debate about whether this organization isa cause or consequence of transcription. Here we induced looping out from chromosome territories (CTs), by the acti-vation of Hox loci during differentiation, to investigate consequences on neighboring loci. We show that, even thoughflanking genes are caught up in the wave of nuclear reorganization, there is no effect on their expression. However, there isa differential organization of active and inactive alleles of these genes. Inactive alleles are preferentially retained within theCT, whereas actively transcribing alleles, and those associated with transcription factories, are found both inside andoutside of the territory. We suggest that the alleles relocated further to the exterior of the CT are those that were alreadyactive and already associated with transcription factories before the induction of differentiation. Hence active generegions may loop out from CTs because they are able to, and not because they need to in order to facilitate geneexpression.
[Supplemental material is available online at www.genome.org.]
Gene organization along the primary DNA sequence of mamma-
Gene clusters where intra-CT organization most closely cor-
lian genomes is not random. Genes that, for the most part, are
relates with expression are those that contain functionally related
functionally unrelated, cluster in the genome and selection has
and coregulated genes. When silent, these clusters, which are mainly
acted to prevent these clusters from becoming fragmented during
located inside of CTs, upon induction of expression will relocate
evolution (Singer et al. 2005). The genes in these clusters tend to
more toward the outside of CTs (Volpi et al. 2000; Williams et al.
be expressed at high levels (Caron et al. 2001) and/or in a wide
2002). Examples of such gene clusters are the murine Hoxb and
range of tissues (Lercher et al. 2002). Moreover, reporter genes are
Hoxd loci. CT reorganization accompanies their induction, both
also expressed at higher levels when inserted into these domains
during the differentiation of embryonic stem (ES) cells and
compared with other genomic regions (Gierman et al. 2007). It has
also along the anterior–posterior axis of the developing embryo
been suggested that gene clustering facilitates transcriptional
(Chambeyron and Bickmore 2004; Chambeyron et al. 2005;
activation by creating regions of open decondensed chroma-
Morey et al. 2007). However, the absence of looping out of Hoxd
tin (Sproul et al. 2005). Similarly, colocalization of active ge-
from its CT in the limb bud (Morey et al. 2007), and the ability
nes at focal concentrations of RNA polymerase II (transcription
of a transposed Hoxb1 to induce some looping out in the absence
factories) or splicing factors (splicing speckles) may enhance the
of transcription (Morey et al. 2008), question the absolute re-
efficiency of gene expression (Shopland et al. 2003; Osborne
quirement for this facet of nuclear reorganization in promoting
et al. 2004; Fraser and Bickmore 2007; Brown et al. 2008). In-
gene expression.
deed, nuclear clustering of gene-rich domains, both in cis and in
Paralogous mammalian Hox loci evolved by duplication
trans, has been detected by diverse techniques (Shopland et al.
of an ancestral cluster during vertebrate radiation (Ferrier and
2006; Simonis et al. 2006). Many of these gene-rich domains have
Minguillon 2003). The apparent similarity in the nuclear behavior
also been seen to adopt positions at the edge or outside of chro-
of Hoxb and Hoxd occurs despite the very different genomic con-
mosome territories (CTs)—termed looping out, but regardless
texts within which they are found. Hoxd is within an extensive
of the activity of individual genes within them (Mahy et al. 2002;
gene desert. Aside from the closely linked Mtx2, Evx2, and Lnp,
Brown et al. 2006). Hence, it remains unclear whether there is a
there are no other annotated genes for 600–700 kilobases (kb) 39 or
causal relationship between CT reorganization and transcription.
59 of Hoxd. In contrast, Hoxb is embedded in a gene-dense region,
In one case, an increased frequency of looping out, induced by
flanked by many unrelated genes (Fig. 1). This raises interesting
the insertion of a beta-globin LCR, did accompany increased asso-
questions as to how genomic environments impact the expression
ciation of the flanking genes with transcription factories and an
and nuclear organization of Hox loci and, conversely, how acti-
activation of some gene expression (Noordermeer et al. 2008).
vation and reorganization of these Hox loci affects the flankinggenes and genomic regions.
Using DNA-fluorescence in situ hybridization (FISH), coupled
with gene expression analysis during ES cell differentiation, we
Present addresses: 1Institut Pasteur, Roux 75015, Paris, France;
show that intra-CT reorganization, initiated within Hox clusters,
Institut National de la Recherche Agronomique, 78352 Jouy-en-
Josas Cedex, France.
then spreads out for hundreds of kilobases into adjacent genomic
regions that, in the case of Hoxb, contain many unrelated genes.
E-mail
[email protected]; fax 44-131-467-8456.
We show that the expression of these flanking genes is not affected
Article published online before print. Article and publication date are athttp://www.genome.org/cgi/doi/10.1101/gr.089045.108.
by their being swept up in extensive CT reorganization. Using
19:1184–1194 Ó 2009 by Cold Spring Harbor Laboratory Press; ISSN 1088-9051/09; www.genome.org
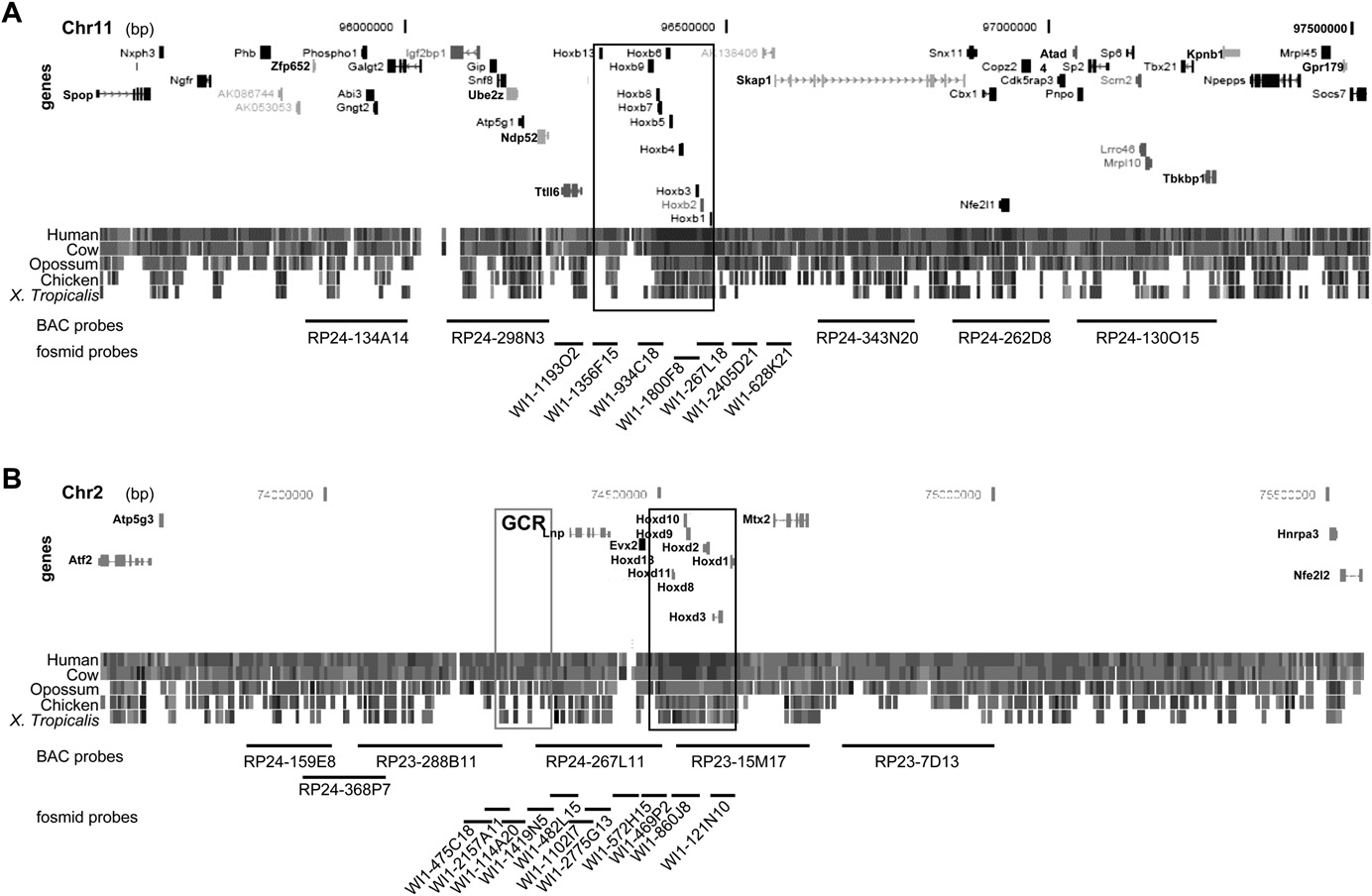
Nuclear reorganization around Hox loci
Comparative genomic structure of Hoxb and Hoxd. Map of the 2 Mb genomic regions surrounding Hoxb on MMU11 (A) and surrounding Hoxd
on MMU2 (B) showing the position of genes, the level of conservation between the murine locus and other vertebrates, and the position of BACs and fosmidsused as probes in this study (Supplemental Table S1). The Hoxb and Hoxd loci are boxed in black. The gray box locates a region of noncoding sequenceconservation that includes the global control region (GCR). Data and map positions (bp) are taken from the August 2005 NCBI Build 35 of the mousegenome (http://genome.ucsc.edu/cgi-bin/hgGateway) and from Ensembl v37, February 2006 (http://www.ensembl.org/Mus_musculus/index.html).
RNA-FISH we show that inactive and active alleles of these flank-
intrinsic to Hox clusters themselves, or whether it is influenced by
ing genes do differ in their intra-CT distribution, with inactive
the flanking genomic regions, we used 2D FISH on nuclei of un-
alleles preferentially retained within the CT. However, the actively
differentiated OS25 ES cells to measure the intra-CT position of
transcribing alleles locate inside, at the edge, and outside of the
signals from probe pairs covering >1 Mb around Hoxb and Hoxd
CT. Similarly, by immuno-FISH we show that the association of
(Figs. 1, 2A; Supplemental Table S1).
gene loci with transcription factories is preferentially, but not ex-
The whole Hoxd region, including flanking gene deserts, lo-
clusively, seen toward the edge and outside of the CT. The alleles
cated well inside the MMU2 CT ($60% alleles at distance >0.4
that are relocated further to the exterior of the CT appear to be
mm) with <15% of alleles found outside (Fig. 2B). A different pic-
those that were already active and that were already associated
ture was seen at Hoxb where, consistent with previous studies
with transcription factories. We suggest that silent Hox loci are
(Chambeyron and Bickmore 2004), significantly more loci (;30%)
restrained to localizations inside of CTs and that, upon their ac-
were seen outside of the respective CT than at Hoxd (P < 10 3). This
tivation, a change in long-range chromatin structure releases this
was especially pronounced at the 59 end of the cluster and extends
constraint. This then allows enough freedom of movement for the
to the 59 flanking region (>40% alleles < 0.2 mm).
chromatin of a large genomic region, which includes not just the
This difference in nuclear organization between Hoxb and
Hox loci but also flanking genes, to be able to now locate in many
Hoxd may reflect the activity of the surrounding regions. To in-
positions relative to the CT.
vestigate this we hybridized cDNA from undifferentiated ES cellsto a mouse 38K cDNA array (GEO accession: GSE15166) and to
a custom tiling array for the Hoxb and Hoxd loci and their sur-rounding regions (Fig. 2C; Supplemental Table S2).
Distinct nuclear behaviors of Hoxb and Hoxd regions
As previously assessed by RT-PCR (Chambeyron and Bickmore
in undifferentiated cells
2004; Chambeyron et al. 2005; Morey et al. 2007), Hoxb and Hoxd
In undifferentiated ES cells, silent Hox clusters are preferen-
genes were expressed either very weakly, or not at all. In contrast,
tially located at the edge or inside of their CTs (Chambeyron and
high steady-state levels of transcripts were detected for some genes
Bickmore 2004; Morey et al. 2007). To determine if this is a feature
59 (Igf2bp1, Snf8, Ube2z, and Ndp52) and 39 (Nfe2l1) of Hoxb.
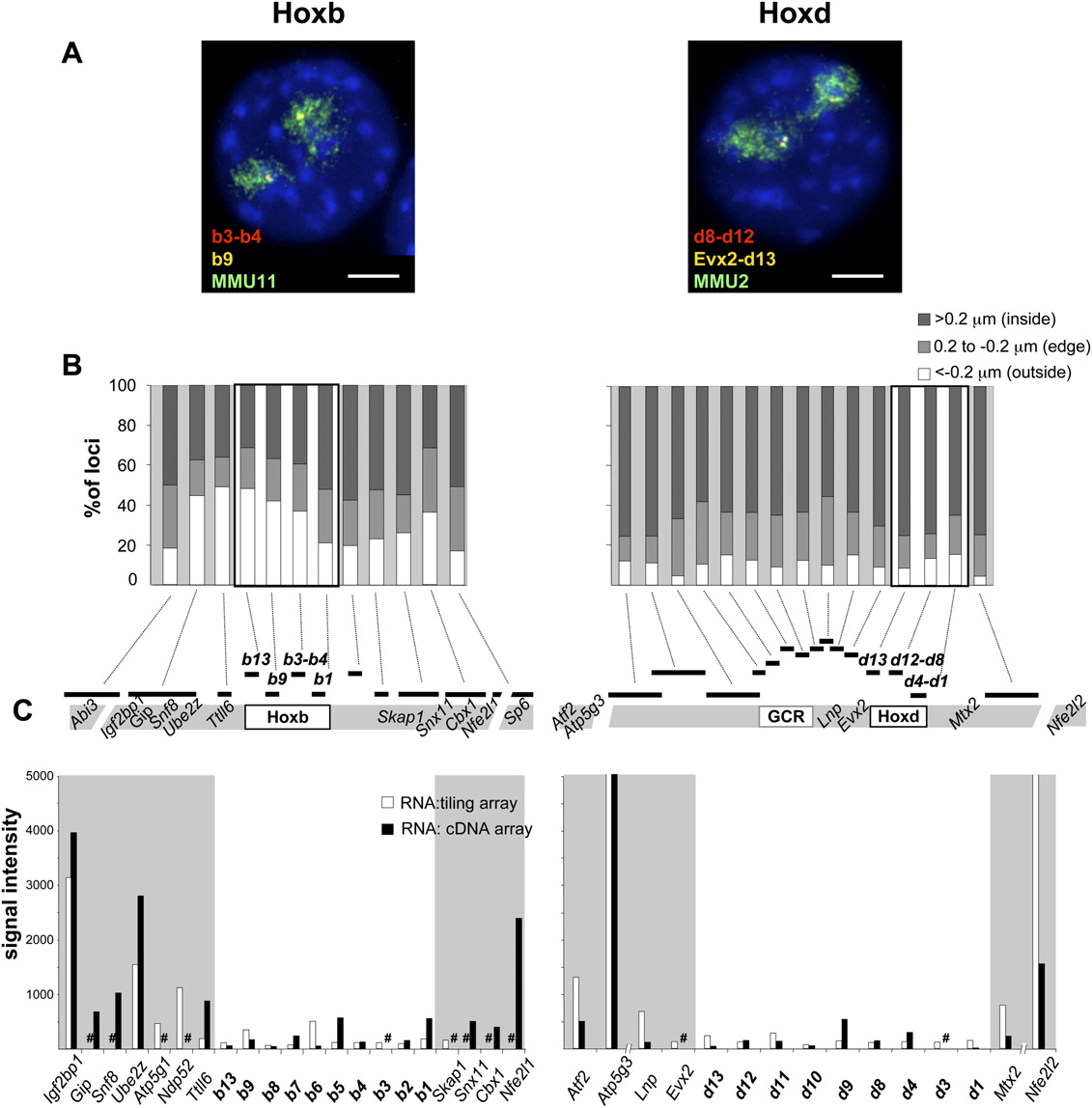
Morey et al.
Nuclear organization and gene expression at Hoxb and Hoxd in undifferentiated ES cells. (A) Four-color DNA-FISH on DAPI counterstained
nuclei from undifferentiated OS25 ES cells using; (left panel) fosmid probes WI1-1800F8 (b3-b4, red), WI1-934C18 (b9, yellow), and an MMU11chromosome paint (green) and (right panel) fosmid probes WI1-860J8 (d8-d12, red), WI1-469P2 (Evx2-d13, yellow), and an MMU2 chromosome paint(green). Bar, 5 mm. (B) Percentage of Hoxb or Hoxd region signals located either inside (>0.2 mm; dark gray bars), at the edge (60.2 mm; light gray bars) oroutside (< 0.2 mm; white bars) of the respective CT edge. The location of the probes is depicted on the map underneath the histograms. A minimum of50 nuclei/100 territories was analyzed. (C ) Intensities of signals of cDNA from undifferentiated OS25 ES cells hybridized on a tiling microarray (whitecolumns) or on a cDNA microarray (black columns). Combined results from three independent experiments, including a dye swap, are shown. A sharpsign indicates genes not represented on the respective array.
Hence, the differential CT organization of Hoxb and Hoxd before
Morey et al. 2007). To determine if this level of nuclear re-
differentiation likely reflects the transcriptional activity of the
organization is restricted just to the Hox genes themselves, or
genes flanking Hoxb.
whether it spreads further, we analyzed the position, relative tothe CT edge, of hybridization signals from probes across the ge-nomic regions, in ES cells that had been differentiated for 3 and 7 d
Chromosome territory reorganization during ES cell
(Fig. 3A,B). As previously reported (Chambeyron and Bickmore
differentiation extends out to the regions that surround
2004), the most extensive looping out was seen at the early stages
of differentiation (D3) and at the 39 end of Hoxb. This decreased by
When activated during ES cell differentiation, an increased fre-
day 7 and, at this time point, looping out had spread to the 59 end
quency of Hox alleles are seen at positions outside of their CTs, as
of Hoxb. A significant relocalization further to the outside of the
measured either by 2D FISH in differentiating ES cells, or by
CT also spreads >500 kb 39 of Hoxb into the flanking gene regions.
3D FISH in differentiating ES cells and in the embryo proper
No additional CT reorganization was detected for the 59 flanking
(Chambeyron and Bickmore 2004; Chambeyron et al. 2005;
domain, which is already mainly located outside, or at the edge, of
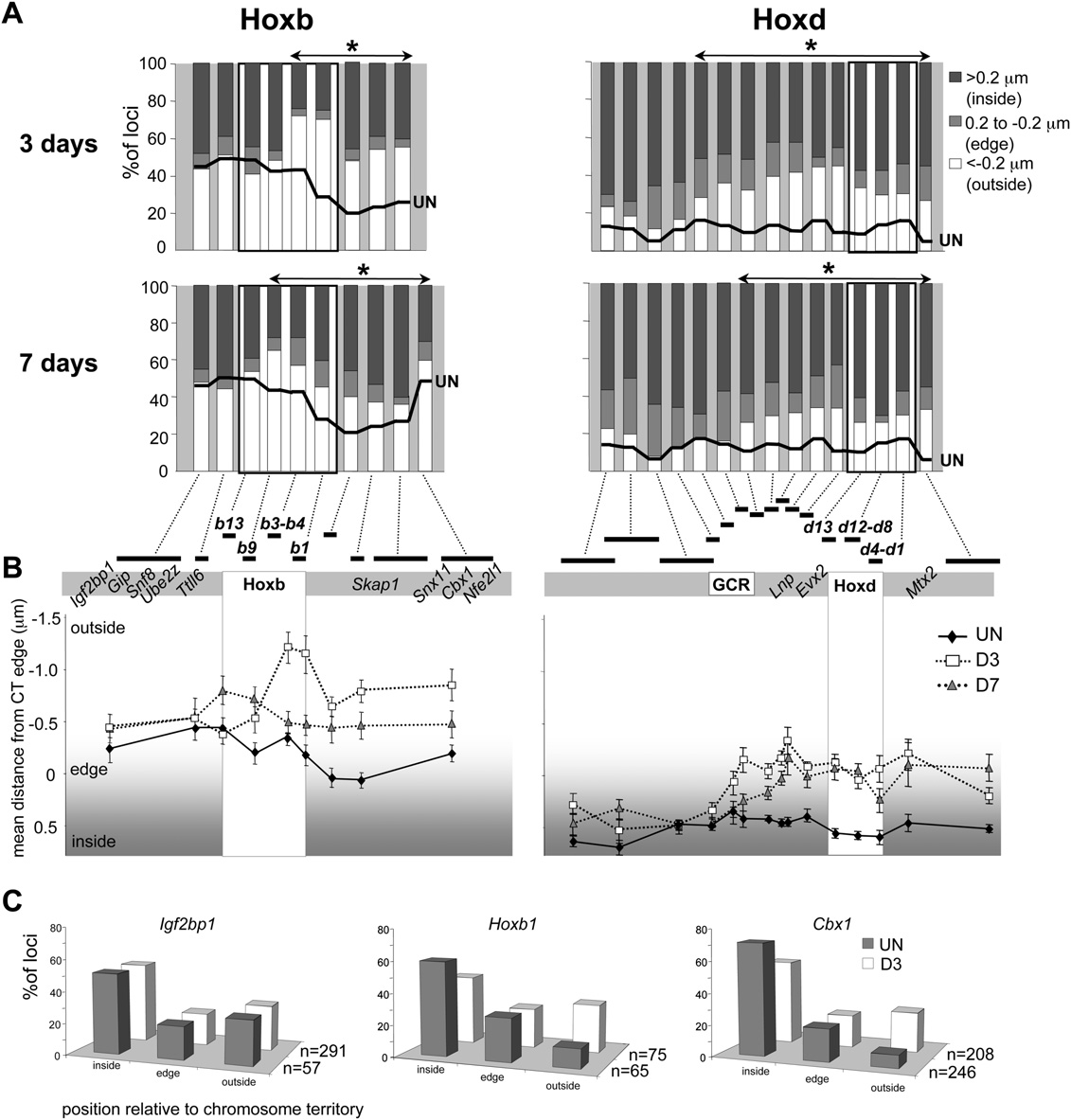
Nuclear reorganization around Hox loci
the CT in undifferentiated cells. This was reconfirmed in 3D FISH
was no significant reorganization (P = 0.165) detected with the
of pFa-fixed cells (Fig. 3C). However, the absolute percentages of
next clone—WI1-475C18. Intriguingly, this boundary to the
Hoxb1, Igf2bp1, or Cbx1 signals located outside, or at the edge, of
spread of CT reorganization 59 of Hoxd corresponds to the end of
the CT during differentiation were lower than those measured by
a long-range regulatory region termed the GCR. As at Hoxb, the
2D FISH. This suggests that, as we have previously described
extent of intra-CT reorganization of Hoxd, decreased by D7.
(Morey et al. 2007), methanol/acetic acid fixation of 2D FISHexaggerates the nuclear movements compared to pFa fixation.
The spread of CT reorganization does not affect expression
At Hoxd, CT reorganization also spreads 39 over Mtx2 and 59
of flanking genes
over Lnp (Fig. 3A,B; Morey et al. 2007). In the 59 direction, FISHsignals from all fosmids from WI1-1102I7 through to WI1-
To test whether the spreading of CT reorganization to the genomic
2157A11 (Fig. 1) showed significant movement toward the edge of
regions flanking Hox clusters impacts the expression of genes lo-
the CT (P < 0.05) at D3 of differentiation (Fig. 3). However, there
cated there, we analyzed gene expression after D3 of differentiation
Chromosome territory reorganization during ES cell differentiation. (A) Histograms showing the percentage of signals across the Hoxb or
Hoxd regions located on either inside (>0.2 mm; dark gray bars), at the edge (60.2 mm; light gray bars), or outside (< 0.2 mm white bars) of the MMU11(Hoxb) or the MMU2 CT (Hoxd), as measured by 2D FISH in OS25 ES cells differentiated for 3 or 7 d. The thick black line shows the corresponding data inundifferentiated ES cells (from Fig. 2B). Probes showing a further significant (P < 0.05) relocalization toward the outside of the CT during differentiationare indicated by the asterisked regions (n = 100). (B ) Mean position (mm) 6 SEM, measured by 2D FISH, of the Hoxb genomic region relative to the edgeof MMU11 CT (left), or of the Hoxd genomic region relative to the edge of MMU2 CT (right) in undifferentiated OS25 ES cells (filled diamonds) and in cellsdifferentiated for 3 d (open squares) or 7 d (shaded triangles). (C ) Position of Hoxb1 and 59 (Ifg2bp1) or 39 (Cbx1) flanking regions relative to the MMU11CT (inside of, edge, or outside of) assayed by 3D FISH in pFa fixed undifferentiated ES cells (filled bars), and cells differentiated for 3 d (open bars).
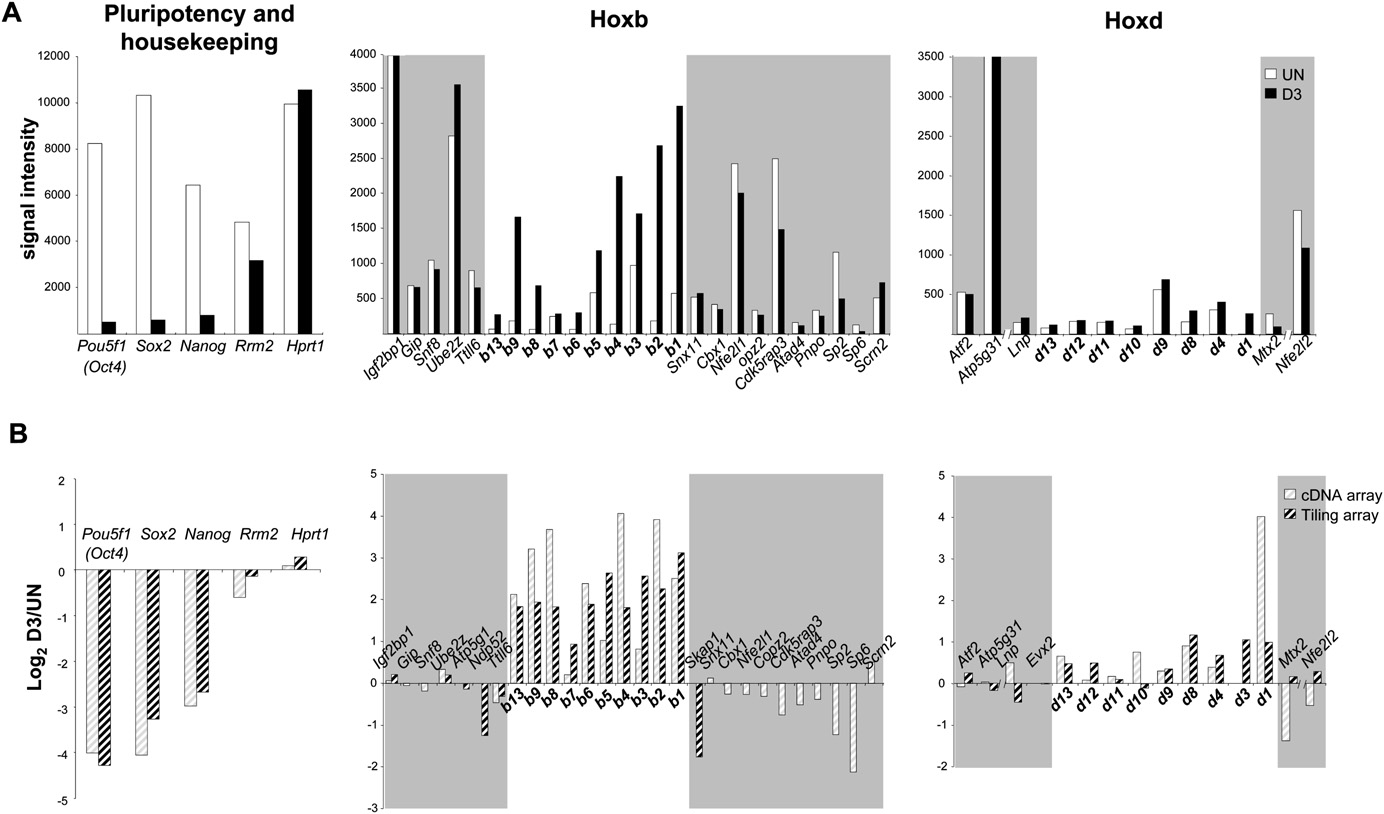
Morey et al.
(Fig. 4). Differentiation was apparent from the dramatically re-
calized to a specific nuclear site to enable their coordinate regu-
duced steady-state levels of transcripts from three markers of
lation by shared regulatory complexes. There is conflicting data
pluripotency (Oct4, Sox2, and Nanog). mRNA levels of two con-
with regard to nuclear colocalization of Hox loci in Drosophila and
stitutively expressed housekeeping genes, Rrm2 and Hprt1, re-
whether this is related to their regulation by polycomb (PcG)
mained constant. There was strong induction of Hoxb genes
complexes (Bantignies et al. 2003; Fedorova et al. 2008). Therefore,
during differentiation (Fig. 4B), and this was supported by RT-PCR
we analyzed the spatial proximity of Hoxb1 and Hoxd1 alleles in ES
data (not shown). In contrast, there was no detectable induction of
cells, before and after differentiation (Fig. 5A). Consistent with
neighboring genes on either side of Hoxb. Indeed, expression of
another study (Lanctot et al. 2007), we did not detect evidence for
Skap1, located immediately 39 of Hoxb1, appeared to be decreased
substantial colocalization, or close association, of Hox loci in trans,
upon differentiation as judged from the tiling array (this gene is
either in the silent (undifferentiated) or active (differentiated)
not present on the expression array used), even though this gene is
state by 2D FISH (Fig. 5B). There was also no colocalization seen by
significantly relocalized further toward the outside of the CT
3D FISH of differentiated cells (Fig. 5C).
during differentiation (Fig. 3). In comparison with Hoxb, Hoxdgenes are only moderately up-regulated during differentiation.
Preferential CT organization of nonexpressing vs. expressing
This might be linked to the differentiation pathways the cells have
engaged in, or the different genomic contexts of the two Hoxclusters.
To more directly address the relationship between gene expression
We conclude that there is no bystander activation of flanking
and CT organization we determined the nuclear distribution of
unrelated genes during Hox gene induction, and that the en-
transcribing alleles using RNA-FISH with intron probes (Supple-
hanced looping out from the CT during Hox activation does not,
mental Table S3) followed by DNA-FISH with genomic probes. We
de facto, lead to the activation of otherwise silent genes, nor does
were unable to visualize nascent RNA signals for Hoxb1 or Hoxd1,
it further enhance the transcription of already active genes.
probably due to the short and single intron structure of theirtranscription units, thus ensuring rapid mRNA processing andexport to the cytoplasm. Therefore, we analyzed the nuclear be-
Hoxb and Hoxd are not colocalized in the nucleus
havior of two constitutively expressed genes, Igf2bp1 and Cbx1,
What then is the functional significance of looping out for the Hox
which flank Hoxb to the 59 and 39 sides, respectively (Fig. 1).
genes themselves? One possibility is that Hox loci are being relo-
Igf2bp1 is in a region that is already substantially (35% of alleles)
Gene expression around Hoxb and Hoxd during ES cell differentiation. (A) Signal intensities of cDNAs from undifferentiated ES cells (UN, white
columns) or from cells differentiated for 3 d (D3, black columns) monitored using a cDNA microarray (GEO accession GSE15166). Results for pluripotency(Oct4, Sox2, Nanog) and housekeeping genes (Rrm2, Hprt1) (left), and the Hoxb (middle) or Hoxd (right) genomic regions are shown. (B) As in A, butshowing the log2 ratio of D3/UN cDNA on a cDNA microarray (light hatched columns) or a Hoxb/Hoxd tiling microarray (dark hatched columns). Data arethe mean of three independent experiments including a dye swap.
Nuclear reorganization around Hox loci
change in the intra-CT distribution of thenontranscribing alleles (P > 0.05). There-fore, it is the already active alleles thatseem to be the ones that can relocatemore to the outside of the CT duringdifferentiation (P = 0.003).
Looping out of Hoxb1, but notof flanking genes, coincides withcolocalization with transcriptionfactories
Nuclear movements are thought to allowfor, or to be a consequence of, recruit-ment of genes to specialized nuclear com-partments. Actively transcribing geneshave been reported to associate with fociof RNA polymerase II (RNAPII)—termedtranscription factories (Osborne et al.
2004). Using immuno-DNA-FISH withan antibody (H5) specific for the Ser2phosphorylated (elongating) form ofRNAPII (RNAPIIo), we analyzed the as-sociation of Igf2bp1, Cbx1, and Hoxb1 ge-nomic regions with these foci (Fig. 7A).
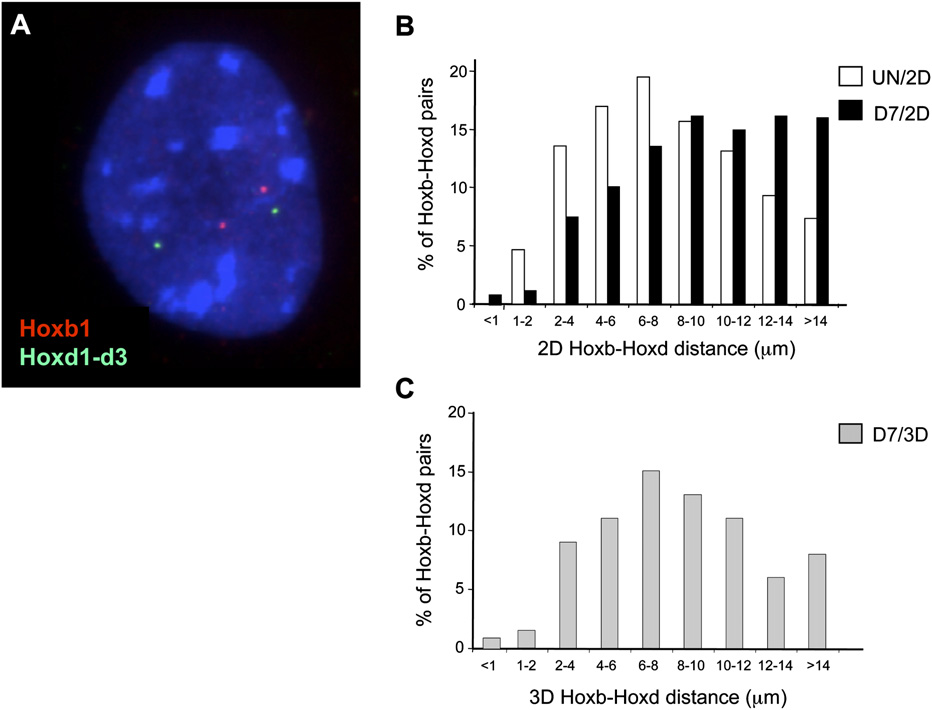
Spatial proximity between Hoxb1 and Hoxd1 loci. (A) Three-color DNA-FISH on DAPI
We quantified the proportion of DNA-
counterstained nucleus from a differentiated (7 d) OS25 ES cell using probe for Hoxb1 (BAC MMP-4; as
FISH signals colocalized with, or dis-
in Chambeyron and Bickmore [2004]) in red, and fosmid probe for Hoxd1-d3 (WI1-121N10) in green.
tinct from, the foci of RNAPIIo (Fig. 7B).
(B) Histogram showing the distribution of distances between the Hoxb and Hoxd signals measured by
Approximately 30% of Cbx1 BAC signals
2D FISH in nuclei from undifferentiated and differentiated (7 d) cells (59 and 87 nuclei analyzed, re-spectively). Distances are slightly larger in differentiated cells due to the increased nuclear volume. (C )
colocalized with RNAPIIo foci, similar to
As in B but using 3D FISH on pFa fixed cells.
the percentage of actively transcribedCbx1 alleles detected in RNA-FISH. Forthe BAC encompassing Igf2bp1 and
localized outside of the CT in undifferentiated cells and this is not
neighboring genes, the percentage of colocalized loci (48%)
further increased upon differentiation (Fig. 3C). In contrast, few
in undifferentiated cells was lower than the proportion of
(8%) Cbx1 signals were seen outside of the CT in undifferentiated
Igf2bp1 alleles with RNA-FISH signals and we do not know the
cells but localization to positions exterior of the CT is increased (to
reason for this. In stark contrast, there was no colocalization be-
>25% of alleles) during differentiation by the spread of CT re-
tween Hoxb1 alleles and RNAPIIo foci in undifferentiated cells
organization emanating from Hoxb (Fig. 3C).
where this Hoxb1 is silent, but in differentiated cells 35% of
In agreement with microarray analysis, expression of Igf2bp1
alleles could now be seen to be colocalized. These data pro-
was more abundant (72% of cells showed an RNA-FISH signal)
vide further support to the idea that much gene transcription
than that of Cbx1 (27%) and this did not significantly change
takes place in association with transcription factories containing
during differentiation (Fig. 6A). We then used DNA-FISH, with
hyperphosphorylated RNAPII (Kimura et al. 2002; Osborne et al.
a MMU11 chromosome paint and a corresponding BAC probe, to
assess the relative CT organization of transcribing vs. nontran-
By comparing the spatial organization of RNAPIIo-colocalized
scribing alleles in these same cells. Analysis was confined to cells
and noncolocalized alleles, we then asked where this recruitment
where RNA-FISH signal was detectable at at least one of the alleles.
to RNAPII foci occurs in relation to CTs (Fig. 7C). Consistent with
Of the nontranscribing (RNA-FISH signal negative) alleles, >72%
the RNA-FISH data (Fig. 6), Cbx1 and Igf2bp1 loci colocalized with
were located well inside of, and <9% were scored as exterior to, the
RNAPIIo inside, at the edge, or outside of the CT. However, both
CT in undifferentiated cells (Fig. 6B). Since RNA-FISH is likely not
before and after differentiation, there was a preferential distribu-
100% efficient and the BACs used for DNA-FISH also encompass
tion of the colocalized alleles away from the CT interior and to-
other neighboring genes (Gip, Snf8, Ube2, and Atp5g1 for the
ward the CT edge and exterior compared to the noncolocalized
Igf2bp1 BAC, and Snx11, Nfe2l1, and Copz2 for Cbx1 BAC), we
alleles (P < 0.001 in x2 test). For Hoxb1, there was also a preference
cannot exclude that some of these latter signals are actually ex-
for the associated alleles to be excluded from the interior of the CT
pressing alleles of these other genes.
(P < 0.001).
The distribution of actively transcribing (RNA-FISH +ve)
To analyze directly the association of a single transcrib-
alleles was significantly different from that of the inactive ones
ing gene with transcription factories, we performed RNA-
(P < 0.0001 by x2). Whereas, the nontranscribing alleles were
immuno-FISH using intronic probes for either Igf2bp1 or Cbx1 on
preferentially located inside the CT, actively transcribing alleles
cells grown on slides, using mild conditions of fixation and per-
were distributed between locations inside, at the edge, and outside
meabilization in order to preserve the nuclear architecture (Fig. 8).
of the CT (Fig. 6B). As the proportion of Cbx1 alleles exterior to the
Greater than 60% of transcribing (RNA-FISH positive) alleles of
CT increased during differentiation, there was no significant
Igf2bp1 and Cbx1 completely colocalized with visible foci of
Morey et al.
pression (Cajiao et al. 2004; Ebisuya et al. 2008) but nothing isknown about nuclear organization in these cases.
Recently, the integration of the beta-globin LCR into a region
of the mouse genome containing a high density of genes withwidespread and diverse expression patterns, was shown to increasethe expression of some of these flanking genes in the embry-onic liver—where the endogenous beta-globin locus is active(Noordermeer et al. 2008). The integrated LCR also inducedboth an increased localization of the region to the edge, and tothe outside, of the CT, and a slightly increased association withfoci of RNAPIIo. Therefore in this case, there was a bystander
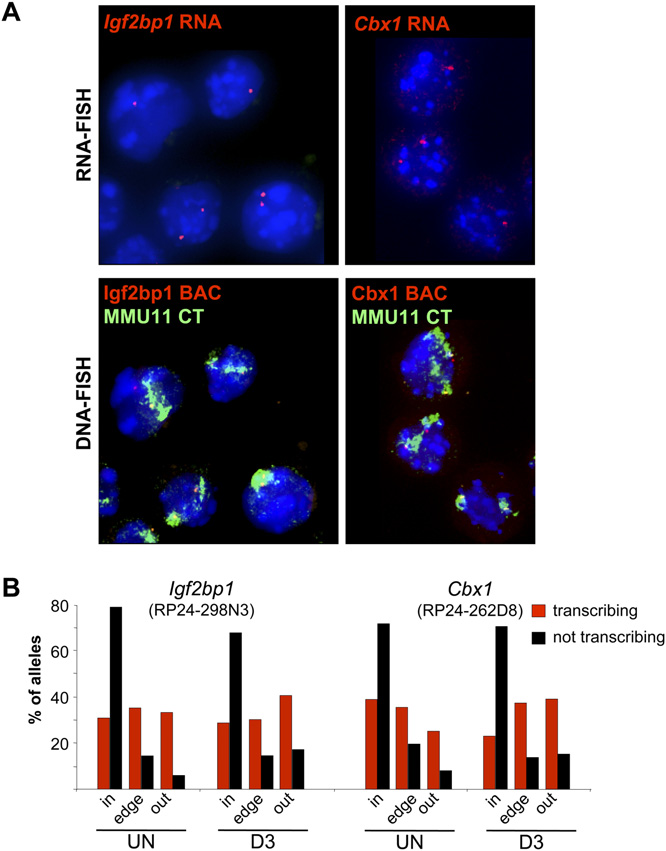
Nuclear organization of actively transcribing alleles. (A) (Top
panels) RNA-FISH with Igf2bp1 or Cbx1 intronic probes (red) on DAPIcounterstained nuclei of cytospun OS25 ES cells. (Lower panels) Samenuclei after denaturation and DNA-FISH with BAC probes overlappingIgf2bp1 (RP24-298N3) and Cbx1 (RP24-262D8) regions (red) togetherwith an MMU11 chromosome paint (green). (B) Histogram showing thepercent of actively transcribing/ RNA-FISH +ve (red) or nontranscribing(black) Igf2bp1 or Cbx1 alleles located inside (in), at the edge (edge), oroutside (out) of the MMU11 CT in undifferentiated (UN) OS25 ES cellsand in cells differentiated for 3 d (D3). Analysis was confined to cells withat least one RNA-FISH signal. For Igf2bp1 n = 288 and 192, UN and D3,respectively. For Cbx1, n = 320 (UN) and 128 (D3).
RNAPIIo and <32% of RNA-FISH negative alleles were colocalized.
To analyze the intra-CT position of active and inactive loci, wethen denatured the nuclei and cohybridized them with a MMU11paint and a BAC probe corresponding to either Cbx1 or Igf2bp1. Asexpected from our previous data, we detected a differential intra-CT localization whether comparing either the RNA-FISH positivevs. negative alleles, or the RNAPIIo colocalized vs. noncolocalizedalleles.
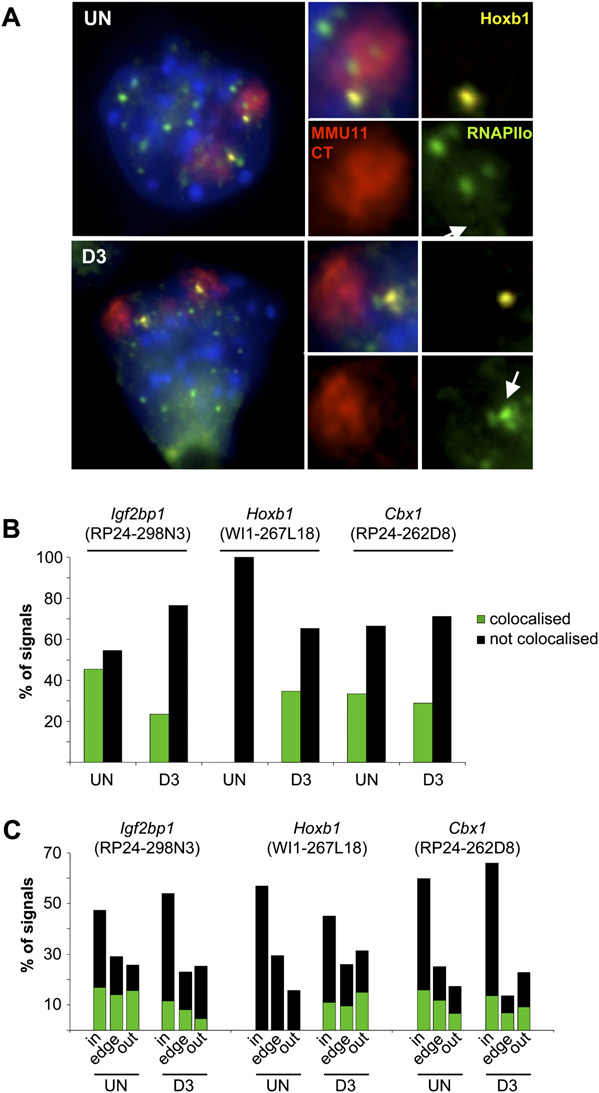
Locus colocalization with foci of RNAPII. (A) Immuno-FISH
using an antibody against the phosphorylated form of RNAPII (green),Hoxb1 probe WI1-267L18 (yellow), and MMU11 chromosome paint (red)
Bystander gene activation
in undifferentiated ES cells (UN) or cells differentiated for 3 d (D3). Theinsets to the right show magnified images from one CT with arrows in-
The clustering of genes in the genome sequence means that
dicating the position of the Hoxb1 locus in the RNAPIIo channel. (B)
nuclear reorganization at one locus will inevitably influence
Histogram showing the percent of signals either colocalized with (dark
the spatial organization of neighboring genes. If nuclear re-
green) or distinct from (not colocalized; black) phosphorylated RNAPII
organization is a directed mechanism for the regulation of gene
foci for the regions encompassing Igf2bp1 (BAC RP24-298N3), Hoxb1(fosmid WI1-267L18) or Cbx1 (BAC RP24-262D8) in undifferentiated
expression, then genes might be activated just because of their
(UN) or differentiated cells (D3). (C ) Histogram of the nuclear distribution
proximity to genes that are subject to nuclear reorganization.
of signals in A with respect to their CT edge. n = 341 (Un), 89 (D3) for
There are documented examples of bystander effects on gene ex-
Igf2bp1; 62 (Un), 73 (D3) for Hoxb1; and 200 (Un), 90 (D3) for Cbx1.
Nuclear reorganization around Hox loci
Preferential localization of inactive alleles insideof chromosome territories
Even though we show that localization of Hox flanking genesfurther to the outside of CTs is not sufficient to increase their ex-pression, we do observe a correlation between gene activity andCT organization. Using combined RNA- and DNA-FISH, we showthat inactive and active alleles have a significantly different dis-tribution within the CT from each other. Although actively tran-scribing alleles, as defined by the presence of a correspondingRNA-FISH signal, are found at positions inside, at the edge, andoutside of the CT, inactive alleles are preferentially retained insideof the CT (Fig. 6).
Actively transcribing genes have been shown to have a pref-
erential association with transcription factories—visibly detect-able concentrations of phosphorylated RNAPII that are coincidentwith sites of BrUTP incorporation (Osborne et al. 2004; Brownet al. 2008). We found that the alleles of Hoxb flanking genes thatcolocalized with RNAPIIo foci were differentially localized in theCT compared to the nonassociated alleles (Fig. 7). The latter arepreferentially found in the CT interior, whereas RNAPIIo colo-calized alleles are distributed between positions inside, at the edge,and outside of the CT. As the frequency of alleles found outside ofthe CT increased during differentiation, there was no further in-crease in transcription factory association, consistent with theabsence of increased gene expression. However, it was the allelesalready associated with transcription factories, and therefore
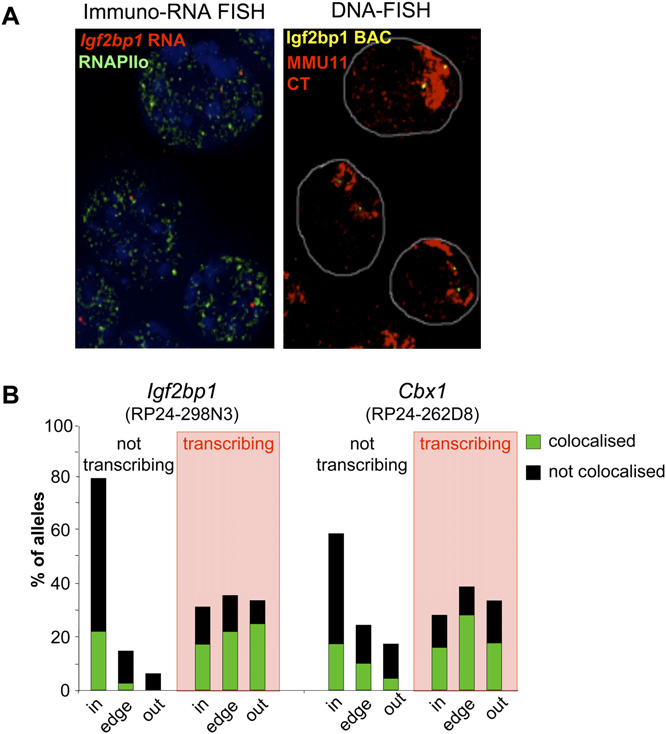
Active vs. inactive allele colocalization with foci of RNAPII. (A)
considered transcriptionally active, that seemed to be the ones
Maximal projection image after deconvolution of 3D RNA-immuno-FISHusing an Igf2bp1 intronic probe (red) and an antibody against the
that preferentially are able to be repositioned in the nucleus.
phosphorylated form of RNAPII (green) on DAPI counterstained nuclei of
Noordermeer et al. (2008) concluded that the increased tran-
undifferentiated OS25 cells (left panel). The DAPI channel has been at-
scription factory association, induced by the beta-globin LCR,
tenuated in order to improve the visualization of red and green signals.
occurred independent of position within the CT. However, in that
The right panel shows the same nuclei after denaturation and DNA-FISHwith a BAC probe encompassing Igf2bp1 (RP24-298N3, yellow) together
study the frequency of alleles scored well inside of the CT, was too
with an MMU11 chromosome paint (red). (B) Histogram showing the
low to exclude that there may be a deficiency of factory-associated
percent of nontranscribing and transcribing (shaded in red) Igf2bp1 or
alleles in the CT interior.
Cbx1 alleles located inside (in), at the edge (edge), or outside (out) of the
Our data are consistent with previous observations of sites of
MMU11 CT together with the percent of alleles colocalized with (green)or distinct from (not associated, black) a phosphorylated RNAPII focus
BrUTP incorporation (Abranches et al. 1998; Verschure et al. 1999)
within each category in undifferentiated OS25 cells. n = 288 (not tran-
or RNAPIIo concentration (Branco and Pombo 2006) within CTs.
scribing), 206 (transcribing) for Igf2bp1 and 128 (not transcribing), and
Together these data suggest that genes in the ‘‘mundane'' genomic
57 (transcribing) for Cbx1.
regions that flank Hox loci, and perhaps most genes in the ge-nome, are transcribed in association with transcription factories(Osborne et al. 2004) that are not restricted to sites at the edge oroutside of the CT. Our data do not distinguish whether genes
effect on both gene expression and CT organization induced by
move to preexisting factories upon activation, or whether factories
might be assembled de novo upon activated genes.
Here we show that there is also a bystander effect on the
We have shown here that Hox gene activation during the
nuclear organization of genes flanking the 39 side of Hoxb. They
differentiation of ES cells, correlates with the association of Hox
increase their localization to the outside of the CT during differ-
loci with transcription factories marked by RNAPII phosphory-
entiation, as a consequence of Hoxb activation and reorganization
lated on Ser2 of the CTD. This is considered to be the actively
(Fig. 3). However, there is no accompanying change in their ex-
elongating form of polymerase. The recent identification of forms
pression (Fig. 4).
of RNAPII phosphorylated on Ser5 at genes, including Hox, si-
It was already known that localization outside of CTs is not
lenced by polycomb in ES cells (Stock et al. 2007), raises the pos-
sufficient to activate genes from an inactive state, but this could be
sibility that in undifferentiated ES cells Hox loci may be associated
attributed to lack of appropriate transcription factors (Mahy et al.
with foci containing this form of RNAPII.
2002; Brown et al. 2006). In our study here, the Hoxb flankinggenes are already active, but we do not observe any further in-
Altered chromatin mobility may explain the link between
crease in their expression as a result of further CT looping out. This
the ability to locate outside of CTs and transcription
is consistent with the conclusion of Noordermeer et al. (2008),which is that looping out is a by-product of LCR activity and that
We conclude that it is not looping out from the CT per se, but
the bystander gene activation is due to direct contacts between
rather the ability of a genome region to be seen as looped out from
this strong regulatory element and flanking genes. There is no
the CT, that is important for active transcription. Looping out
known long-range regulatory element needed for Hoxb activation
from the CT is therefore likely to be a manifestation of some
during ES cell differentiation.
other underlying change in chromatin structure that enhances
Morey et al.
chromatin mobility (Fraser and Bickmore 2007). This might then
rectly. One microliter of RNase inhibitor (Invitrogen) was added
allow genes the freedom to explore a larger proportion of the
to the hybridization mix. Hybridization conditions were the same
nuclear space, both inside and outside of the CT, and hence
as for DNA-FISH. Slides were washed in 50% formamide 23 SSC
increases the probability that a gene will be able to engage with
(pH 7.5) at room temperature, then at 37°C and then in 23 SSC at
a transcription factory. In this model, it is the restraint on chro-
room temperature. Signal detection was as for DNA-FISH. A con-
matin motion, for alleles that are not able to move outside of CTs,
trol with treatment with RNase confirmed signal specificity. A
that restricts their ability to be transcribed and not their intra-CT
minimum of 50 2D images comprising at least one nucleus with
position per se. This then raises the challenge of identifying the
an RNA-FISH signal were captured and the slide coordinatesrecorded. Standard DNA-FISH was then applied and the same cells
chromatin structures that modulate chromatin mobility.
relocated on the slide.
ES cell culture and differentiation
Immunofluorescence followed by DNA-FISH was as described athttp://www.epigenome-noe.net/researchtools/protocol.php?protid=
OS25 ES cells were cultured as previously described (Chambeyron
3 (Chaumeil et al. 2004). Cells grown on slides were fixed with 4%
and Bickmore 2004; Morey et al. 2007). Differentiation was in-
pFa (pH 7.4) for 15 min at room temperature, washed twice in ice-
duced by plating the cells at low density without LIF or hygro-
cold PBS, and stored at 4°C in PBS. Before immunostaining, cells
mycin for 1 d. Retinoic acid (RA), 5 3 10 6 M, was then added.
were permeabilized with 0.5% Triton X-100/PBS for 12 min at room
RNA samples were collected and nuclei were prepared after 3 or
temperature. Cells were then incubated with the anti-RNAPII H5
7 d of differentiation (respectively, 2 d of RA treatment and
(1/100, Covance) and detected with a fluorescein anti-mouse (1/100,
4 d with and 2 d without RA). Similar FISH results were obtained
Vector Laboratories) secondary antibody. Slides were post-fixed with
on samples collected from two independent differentiation
3% formaldehyde for 10 min at room temperature, incubated in 0.1
experiments. FISH data presented in this paper all come from the
M HCl for 10 min at room temperature, denatured for 30 min at
80°C in 50% formamide 23 SSC (pH 7.5), and hybridized.
Multicolor DNA fluorescence in situ hybridization
For 2D FISH, nuclei were isolated in 0.56% KCl and fixed with
Cells grown on slides were fixed with 3% pFA (pH 7.4) for 10 min
3:1 v/v methanol/acetic acid (Chambeyron and Bickmore 2004).
on ice, permeabilized with 0.5% Triton X-100/PBS complemented
For 3D FISH, nuclei were fixed with 4% paraformaldehyde (pFa).
with 2 mM VRC for 5 min on ice, washed once in 23 SSC, hy-
FITC-labeled paint for MMU2 or MMU11 were purchased from
bridized overnight, and washed as described above. Slides were
CAMBIO. BAC and fosmid clones covering the Hoxb and Hoxd
post-fixed with 3% pFa for 10 min at room temperature and
regions were chosen from Ensembl v37, February 2006 (http://
repermeabilized with 0.5% Triton X-100/PBS complemented with
www.ensembl.org/Mus_musculus/index.html). BAC clones were
2 mM VRC for 5 min on ice. Immunostaining procedure was
purchased from BACPAC resources center and fosmid clones were
as described above. One-one hundreths of RNase inhibitor was
kindly provided by the Sanger Institute (for coordinates and
added to blocking buffers and primary and secondary antibodies.
names, see Supplemental Table S1). DNA from these clones were
Three-dimensional images of nuclei with at least one RNA-FISH
prepared using standard alkaline lysis and labeled with digox-
signal were captured and the coordinates of these cells recorded
igenin-11-dUTP or with biotin-16-dUTP (Roche) by nick trans-
before DNA-FISH.
lation. Approximately 200 ng of FITC-paint, 100 ng of biotin-labeled BAC/fosmid probe, and 100 ng of digoxigenin BAC/fosmid
Image capture and analysis
probe were used per slide, together with 15mg of mouse Cot-1 DNA
Two-dimensional slides were examined using a Zeiss Axioplan II
(GIBCO BRL) and 5 mg salmon sperm DNA. Digoxigenin-labeled
fluorescence microscope with Plan-neofluar objectives, a 100 W
probes were detected using Rhodamine anti-digoxigenin and
Hg source (Carl Zeiss) and Chroma #84000 quadruple band pass
Texas red anti-sheep IgG (Vector Laboratories); biotin-labeled
filter set (Chroma Technology Corp.) with the excitation filters
probes were detected using Cy5 streptavidin and biotinylated anti-
installed in a motorized filter wheel (Ludl Electronic Products).
avidin (Vector Laboratories) and the FITC signal from the MMU2
Grayscale images were captured with a Hamamatsu Orca AG CCD
or MMU11 chromosome paint was amplified using F1 rabbit anti-
camera (Hamamatsu Photonics Ltd.). Image capture and analysis
FITC and F2 FITC anti-rabbit (CAMBIO). Hybridization, washes,
were performed using in-house scripts written for IPLab Spectrum
detection, and imaging were as described (Chambeyron and
(Scanalytics Corp.). The distance between two probes, and probe
Bickmore 2004). Slides were mounted in Vectashield (Vector) and
position relative to the CT edge, were calculated as described
counterstained with 0.5 mg/mL DAPI.
(Chambeyron and Bickmore 2004; Chambeyron et al. 2005;Morey et al. 2007). In this analysis a probe signal is considered
to be outside of the CT when its distance to the CT edge is nega-tive. Conversely, distances >0 correspond to probe signals inside
Probes for RNA-FISH consisted of a mix of PCR products from the
introns of Igf2bp1 or Cbx1 and were labeled using nick translation.
Three-dimensional images were captured on a Zeiss Axioplan
Primer sequences are shown in Supplemental Table S3. The RNA-
II fluorescence microscope with the objective fitted with a Pifoc
FISH procedure was as previously described (Debrand et al. 1999).
motor. Single plane images were deconvolved using the iterative
Briefly, cells were cytospun onto SUPERFROST glass slides (4 min,
restoration (20 iterations) from Volocity (Improvision) software.
350 rpm, Shandon Cytospin), permeabilized with 0.5% TritonX-100 in ice-cold cytoskeletal buffer complemented with 2 mM
Statistical analysis
Vanadyl Ribonucleoside Complex (VRC) (New England Biolabs)for 7 min, fixed with 4% pFa on ice for 10 min and stored in 70%
The statistical relevance of DNA-FISH CT position data was as-
ethanol at 4°C. Slides were then dehydrated and hybridized di-
sessed using the nonparametric Kolmogorov–Smirnov test to
Nuclear reorganization around Hox loci
examine the null hypothesis that two sets of data show the same
slide printing, and James Prendergast (MRC HGU) for Lowess
distribution. The data sets consisted of at least 50 nuclei (100
normalization of data.
territories/loci) for each differentiation time and for each combi-natory of probes. A P-value < 0.05 was considered statisticallysignificant.
The statistical significance of RNA/DNA-FISH and immuno-
DNA-FISH data was assessed by x2.
Abranches R, Beven AF, Ragon-Alcaide L, Shaw PJ. 1998. Transcription sites
are not correlated with chromosome territories in wheat nuclei. J CellBiol 143: 5–12.
Bantignies F, Grimaud C, Lavrov S, Gabut M, Cavalli G. 2003. Inheritance of
Microarray design and hybridization
Polycomb-dependent chromosomal interactions in Drosophila. Genes &Dev 17: 2406–2420.
The mouse cDNA oligonucleotide microarray represented 38,000
Branco MR, Pombo A. 2006. Intermingling of chromosome territories in
mouse genes from the Mouse Exonic Evidence Based Oligonucle-
interphase suggests role in translocations and transcription-dependentassociations. PLoS Biol 4: e138. doi: 10.1371/journal.pbio.0040138.
otide set (van den Ijssel et al. 2005) (GEO accession: GPL4468). For
Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs
the custom tiling array, 60-mer oligos (Invitrogen/Illumina) were
DR, Iborra FJ, Buckle VJ. 2006. Coregulated human globin genes
designed alternatively on each strand of mouse chromosome 2
are frequently in spatial proximity when active. J Cell Biol 172: 177–
and 11 to cover Hoxd and Hoxb core regions, proximal flanking
Brown JM, Green J, das Neves RP, Wallace HA, Smith AJ, Hughes J, Gray N,
regions, and distal flaking regions (one oligo every 250, 500, and
Taylor S, Wood WG, Higgs DR, et al. 2008. Association between active
50 kb, respectively). Additional oligos were designed on exons of
genes occurs at nuclear speckles and is modulated by chromatin
genes of interest: Hoxb, Hoxd, flanking genes, differentiation-
environment. J Cell Biol 182: 1083–1097.
related genes, and housekeeping genes (Supplemental Table S2).
Cajiao I, Zhang A, Yoo EJ, Cooke NE, Liebhaber SA. 2004. Bystander gene
activation by a locus control region. EMBO J 23: 3854–3863.
Amine-modified oligos were spotted at 20 mM in 13 Nexterion
Caron H, van Schaik B, van der Mee M, Baas F, Riggins G, van Sluis P,
Spot buffer containing 0.005% Triton X100 onto Codelink acti-
Hermus MC, van Asperen R, Boon K, Voute PA, et al. 2001. The human
vated slides by the University of Liverpool Microarray Facility.
transcriptome map: Clustering of highly expressed genes in
Total RNA was extracted from OS25 cells with TriReagent
chromosomal domains. Science 291: 1289–1292.
Chambeyron S, Bickmore WA. 2004. Chromatin decondensation and
(Sigma). Ten micrograms of RNA were reverse-transcribed using
nuclear reorganization of the HoxB locus upon induction of
SuperScript Direct cDNA labeling kit (Invitrogen) to produce oligo-
transcription. Genes & Dev 18: 1119–1130.
dT primed cDNAs labeled with Cy3 or Cy5-dCTP (Amersham).
Chambeyron S, Da Silva NR, Lawson KA, Bickmore WA. 2005. Nuclear re-
cDNAs were purified with a Qiagen PCR purification kit and
organisation of the Hoxb complex during mouse embryonicdevelopment. Development 132: 2215–2223.
quantified with a Nanodrop ND-1000 spectrophotometer. Equiv-
Chaumeil J, Okamoto I, Heard E. 2004. X chromosome inactivation in
alent amounts of cDNAs (1–2 mg cDNA corresponding to about
mouse embryonic stem cells: Analysis of histone modifications and
100 pmol of incorporated dye) from undifferentiated and differ-
transcriptional activity using immunofluorescence and FISH. Methods
entiated cells labeled with a different dye were precipitated to-
Enzymol 376: 405–419.
Debrand E, Chureau C, Arnaud D, Avner P, Heard E. 1999. Functional
gether with 10 mg of salmon sperm DNA, 500 mg of yeast tRNA and
analysis of the DXPas34 locus, a 39 regulator of Xist expression. Mol Cell
resuspended in 45 mL of hybridization buffer (50% deionized
Biol 19: 8513–8525.
formamide, 53 SSC, 53 Denhardt, 0.1% SDS). This hybridization
Ebisuya M, Yamamoto T, Nakajima M, Nishida E. 2008. Ripples from
mix was denaturated for 8 min at 94°C and pipetted onto the
neighbouring transcription. Nat Cell Biol 10: 1106–1113.
Fedorova E, Sadoni N, Dahlsveen IK, Koch J, Kremmer E, Eick D, Paro R,
prewarmed microarray slide. Hybridization was overnight under
Zink D. 2008. The nuclear organization of Polycomb/Trithorax group
a 22 3 64 mm coverslip in a humid chamber floating in a 42°C
response elements in larval tissues of Drosophila melanogaster.
water bath. Slides were washed with gentle shaking as follows: 23
Chromosome Res 16: 649–673.
5 min in 23 SSC, 0.1% SDS at 42°C; 23 5 min in 0.23 SSC at room
Ferrier DE, Minguillon C. 2003. Evolution of the Hox/ParaHox gene clusters.
Int J Dev Biol 47: 605–611.
temperature; 23 5 min in 0.13 SSC at room temperature; and 30 s
Fraser P, Bickmore W. 2007. Nuclear organization of the genome and the
in 0.013 SSC at room temperature. Slides were dried by centrifu-
potential for gene regulation. Nature 447: 413–417.
gation at 1000 rpm, 5 min at room temperature.
Gierman, HJ, Indemans MH, Koster J, Goetze S, Seppen J, Geerts D, van
Slides were scanned using Genepix (Axon Instruments).
Driel R, Versteeg R. 2007. Domain-wide regulation of gene expression inthe human genome. Genome Res 17: 1286–1295.
Analysis was done with BlueFuse software (Cambridge Blue-
Kimura H, Sugaya K, Cook PR. 2002. The transcription cycle of RNA
Gnome). Normalization between the two channels (i.e., undif-
polymerase II in living cells. J Cell Biol 159: 777–782.
ferentiated/differentiated cells cDNAs) was by global Lowess for
Lanctot C, Kaspar C, Cremer T. 2007. Positioning of the mouse Hox gene
the cDNA library microarrays, and by global median based on
clusters in the nuclei of developing embryos and differentiatingembryoid bodies. Exp Cell Res 313: 1449–1459.
housekeeping genes for the Hox tiling arrays. For the tiling arrays
Lercher MJ, Urrutia AO, Hurst LD. 2002. Clustering of housekeeping genes
there were two biological and one technical (dye-swap) replicates,
provides a unified model of gene order in the human genome. Nat Genet
and for the expression arrays there were two replicates. These were
31: 180–183.
normalized to each other by a Lowess normalization.
Mahy NL, Perry PE, Bickmore WA. 2002. Gene density and transcription
influence the localization of chromatin outside of chromosometerritories detectable by FISH. J Cell Biol 159: 753–763.
Morey C, Da Silva NR, Perry P, Bickmore WA. 2007. Nuclear reorganisation
and chromatin decondensation are conserved, but distinct,mechanisms linked to Hox gene activation. Development 134: 909–
C.M. was an EMBO long-term fellow and then was a recipient of
a Marie Curie Intra European Fellowship (MEIF-CT-2006-021308);
Morey C, Da Silva NR, Kmita M, Duboule D, Bickmore WA. 2008. Ectopic
C.K. was supported by the EU FP6 Network of Excellence Epi-
nuclear reorganisation driven by a Hoxb1 transgene transposed intoHoxd. J Cell Sci 121: 571–577.
genome (LSHG-CT-2004-503433); W.A.B. is a Centennial Fellow of
Noordermeer, D, Branco MR, Splinter E, Klous P, van IJcken W,
the James S. McDonnell foundation. We thank Bauke Ylstra (Micro
Swagemakers S, Koutsourakis M, van der Spek P, Pombo A, de Laat W.
Array Facility, VUMC Cancer Center Amsterdam) for the mouse
2008. Transcription and chromatin organization of a housekeeping
expression arrays, Nick Gilbert (University of Edinburgh Cancer
gene cluster containing an integrated beta-globin locus control region.
PLoS Genet 4: e1000016. doi: 10.1371/journal.pgen.1000016.
Research Centre) for help with Hox tiling microarray design,
Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E,
Margaret Hughes (Liverpool Microarray Facility) for microarray
Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. 2004. Active genes
Morey et al.
dynamically colocalize to shared sites of ongoing transcription. Nat
restrains poised RNA polymerase II at bivalent genes in mouse ES cells.
Genet 36: 1065–1071.
Nat Cell Biol 9: 1428–1435.
Shopland LS, Johnson CV, Byron M, McNeil J, Lawrence JB. 2003.
van den Ijssel, P, Tijssen M, Chin SF, Eijk P, Carvalho B, Hopmans E,
Clustering of multiple specific genes and gene-rich R-bands around SC-
Holstege H, Bangarusamy DK, Jonkers J, Meijer GA, et al. 2005. Human
35 domains: Evidence for local euchromatic neighborhoods. J Cell Biol
and mouse oligonucleotide-based array CGH. Nucleic Acids Res 33: e192.
162: 981–990.
Shopland LS, Lynch CR, Peterson KA, Thornton K, Kepper N, Hase J, Stein S,
Verschure, P.J., van der Kraan I, Manders EM, van Driel R. 1999. Spatial
Vincent S, Molloy KR, Kreth G, et al. 2006. Folding and organization of
relationship between transcription sites and chromosome territories. J
a contiguous chromosome region according to the gene distribution
Cell Biol 147: 13–24.
pattern in primary genomic sequence. J Cell Biol 174: 27–38.
Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, Campbell
Simonis M, Klous P Splinter E, Moshkin Y, Willemsen R, de Wit E, van
RD, Goldsworthy M, Powis SH, Ragoussis J, et al. 2000. Large-scale
Steensel B, de Laat W. 2006. Nuclear organization of active and inactive
chromatin organization of the major histocompatibility complex and
chromatin domains uncovered by chromosome conformation capture-
other regions of human chromosome 6 and its response to interferon in
on-chip (4C). Nat Genet 38: 1348–1354.
interphase nuclei. J Cell Sci 113: 1565–1576.
Singer GA, Lloyd AT, Huminiecki LB, Wolfe KH. 2005. Clusters of co-
Williams RR, Broad S, Sheer D, Ragoussis J. 2002. Subchromosomal
expressed genes in mammalian genomes are conserved by natural
positioning of the epidermal differentiation complex (EDC) in
selection. Mol Biol Evol 22: 767–775.
keratinocyte and lymphoblast interphase nuclei. Exp Cell Res 272: 163–
Sproul D, Gilbert N, Bickmore WA. 2005. The role of chromatin structure
in regulating the expression of clustered genes. Nat Rev Genet 6: 775–781.
Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff
N, Fisher AG, Pombo A. 2007. Ring1-mediated ubiquitination of H2A
Received November 9, 2008; accepted in revised form March 19, 2009.
Source: http://ftp.hgu.mrc.ac.uk/wendy/pdfs/2009/MoreyGR2009l.pdf
URISTAT • If your symptoms persist, are severe, or you experience fever, chills, back pain or bloody urine see your doctor promptly. You may have a serious condition that requires different treatment. • Phenazopyridine HCI may cause gastrointestinal upset in some people. Take with or after meals to reduce gastric upset and Relief PAK discontinue use if symptoms occur.
Contents lists available at Tetrahedron Letters advances in bioorthogonal reactions for site-specific proteinlabeling and engineering Yukang Gong, Lifeng Pan Key Laboratory of Bioorganic and Natural Products Chemistry, Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, Lingling Road 345, Shanghai 200032,People's Republic of China In the past two decades, with the rapid development of chemical biology, tremendous small-molecule








