Levitra enthält Vardenafil, das eine kürzere Wirkdauer als Tadalafil hat, dafür aber schnell einsetzt. Männer, die diskret bestellen möchten, suchen häufig nach levitra kaufen ohne rezept. Dabei spielt die rechtliche Lage in der Schweiz eine wichtige Rolle.
Pp150722 1263.1273
The Stromal Chloroplast Deg7 Protease Participates inthe Repair of Photosystem II after Photoinhibitionin Arabidopsis1[W][OA]
Xuwu Sun, Tingjiao Fu, Ning Chen, Jinkui Guo, Jinfang Ma, Meijuan Zou, Congming Lu, and Lixin Zhang*
Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy ofSciences, Beijing 100093, China
Light is the ultimate source of energy for photosynthesis; however, excessive light leads to photooxidative damage and hencereduced photosynthetic efficiency, especially when combined with other abiotic stresses. Although the photosystem II (PSII)reaction center D1 protein is the primary target of photooxidative damage, other PSII core proteins are also damaged anddegraded. However, it is still largely unknown whether degradation of D1 and other PSII proteins involves previouslyuncharacterized proteases. Here, we show that Deg7 is peripherally associated with the stromal side of the thylakoidmembranes and that Deg7 interacts directly with PSII. Our results show that Deg7 is involved in the primary cleavage ofphotodamaged D1, D2, CP47, and CP43 and that this activity is essential for its function in PSII repair. The double mutants deg5deg7 and deg8 deg7 showed no obvious phenotypic differences under normal growth conditions, but additive effects wereobserved under high light. These results suggest that Deg proteases on both the stromal and luminal sides of the thylakoidmembranes are important for the efficient PSII repair in Arabidopsis (Arabidopsis thaliana).
Chloroplasts of higher plants carry out one of the
mass proteins (Nelson and Yocum, 2006). The PSII
most important biochemical reactions: the capture of
reaction center D1 protein has been identified among
light energy and its conversion into chemical energy.
PSII proteins as the primary target of light-induced
Although light is the ultimate source of energy for
damage (Kyle et al., 1984; Mattoo et al., 1984; Ohad
photosynthesis, it can also be harmful to plants.
et al., 1984; Adir et al., 1990), but several studies have
Light-induced loss of photosynthetic efficiency, which
shown that the D2, CP47, and CP43 proteins are
is generally termed as photoinhibition, limits plant
degraded under photoinhibitory conditions (Schuster
growth and lowers productivity, especially when com-
et al., 1988; Yamamoto and Akasaka, 1995; Jansen et al.,
bined with other abiotic stresses.
1999; Adir et al., 2003). Moreover, several small PSII
The main target of photoinhibition is PSII, which
subunits, such as PsbH, PsbW, and Cyt b
catalyzes the light-dependent water oxidation con-
found to be frequently replaced within PSII (Hagman
comitantly with oxygen production (for review, see
et al., 1997; Ortega et al., 1999; Bergantino et al., 2003).
Prasil et al., 1992; Aro et al., 1993; Adir et al., 2003). In
Evidence for the involvement of two families of pro-
higher plants, PSII consists of more than 20 subunits,
teases, FtsH and Deg, in the degradation of the D1
including the reaction center D1 and D2 proteins,
protein in thylakoids of higher plants has been re-
cytochrome (Cyt) b
, the light-harvesting chlorophyll
cently described (Lindahl et al., 1996, 2000; Bailey
a-binding proteins CP47 and CP43, the oxygen-evolv-
et al., 2002; Sakamoto et al., 2003; Silva et al., 2003;
ing 33-kD protein (PsbO), and several low molecular
Kapri-Pardes et al., 2007; Sun et al., 2007a, 2007b).
However, it is still largely unknown whether degra-
dation of D1 and other PSII proteins involves previ-
This work was supported by the Major State Basic Research
Development Program (grant no. 2009CB118500), the National Nat-
ously uncharacterized proteases.
ural Science Foundation of China (grant nos. 30725003 and
DegP (or HtrA) proteases were initially identified
30870182), and the Frontier Project of Knowledge Innovation Engi-
based on the fact that they are required for the survival
neering of the Chinese Academy of Sciences (grant no. KJCX2–SW–
of Escherichia coli at high temperatures and for the deg-
radation of abnormal periplasmic proteins (Lipinska
* Corresponding author; e-mail
[email protected].
et al., 1988; Strauch and Beckwith, 1988). DegP is an
The author responsible for distribution of materials integral to the
ATP-independent Ser endopeptidase, and it contains a
findings presented in this article in accordance with the policy
trypsin-like protease domain at the N terminus, fol-
described in the Instructions for Authors (www.plantphysiol.org) is:
lowed by two PDZ domains (Gottesman, 1996; Pallen
Lixin Zhang (
[email protected]).
and Wren, 1997; Clausen et al., 2002). PDZ domains
The online version of this article contains Web-only data.
[OA] Open Access articles can be viewed online without a sub-
appear to be important for complex assembly and
substrate binding through three or four residues in
the C terminus of their target proteins (Doyle et al.,
Plant PhysiologyÒ, March 2010, Vol. 152, pp. 1263–1273, www.plantphysiol.org Ó 2010 American Society of Plant Biologists
1996; Harris and Lim, 2001). DegP switches between
may not be involved in D1 degradation in vivo
chaperone and protease functions in a temperature-
(Huesgen et al., 2006).
dependent manner. The chaperone function dominates
Here, we have expressed and purified a recombi-
at low temperatures, and DegP becomes proteolytically
nant DegP protease, His-Deg7. In vitro experiments
active at elevated temperatures (Spiess et al., 1999).
showed that His-Deg7 is proteolytically active toward
Crystal structures of different members of the DegP
the PSII proteins D1, D2, CP43, and CP47. In vivo
protein family (Krojer et al., 2002; Li et al., 2002; Kim
analyses of a deg7 mutant revealed that the mutant is
et al., 2003; Wilken et al., 2004) have revealed the
more sensitive to high light stress than the wild-type
structure-function relationship of these PDZ-containing
plants. We demonstrated that Deg7 is a chloroplast
proteases. Trimeric DegP is the functional unit, and the
stroma protein associated with the thylakoid mem-
hexameric DegP is formed via the staggered association
branes and that it interacts with PSII, which suggests
of trimers (Clausen et al., 2002; Kim and Kim, 2005). At
that it can cleave the stroma-exposed region of sub-
normal growth temperatures, the active site of the
strate proteins. Our results also provide evidence that
protease is located within the chamber of hexameric
Deg7 is important for maintaining PSII function.
DegP, which is not accessible to the substrates. How-ever, at high temperatures, conformational changesinduce the activation of the protease function (Krojeret al., 2002). Recent studies have shed light on the
substrate binding-induced formation of larger oligo-
Deg7 Is a Chloroplast Stromal Protein
meric complexes of DegP (Jiang et al., 2008; Krojer et al.,2008).
Deg7 was predicted to be located in chloroplasts by
In Arabidopsis (Arabidopsis thaliana), 16 genes cod-
LOCtree. To determine the subcellular localization, the
ing for DegP-like proteases have been identified, and
N-terminal 243 amino acids of Deg7 were fused to the
at least seven gene products are predicted to be located
N terminus of the synthetic GFP under the control of
in chloroplasts (Kieselbach and Funk, 2003; Huesgen
the 35S promoter with a S65T mutation. The Deg7-GFP
et al., 2005; Adam et al., 2006; Sakamoto, 2006; Kato
fusion proteins were transiently expressed in proto-
and Sakamoto, 2009). Based on proteomic data, four
plasts, and GFP fluorescence of Deg7-GFP fusion
Deg proteases have been shown to be localized to the
proteins was colocalized with the chloroplastic chlo-
chloroplast (Peltier et al., 2002; Schubert et al., 2002)
rophyll, which is consistent with the signals observed
and functionally characterized. Deg1, Deg5, and Deg8
when the GFP was fused to the transit peptide of
are located in thylakoid lumen, and Deg2 is peripher-
FtsH11 (Sakamoto et al., 2003). When the targeting
ally associated with the stromal side of thylakoid
signals of the fibrillarin and FRO1 proteins from
membranes (Itzhaki et al., 1998; Haußu¨hl et al., 2001;
Arabidopsis were fused to GFP (Cai et al., 2009),
Sun et al., 2007a). Recombinant DegP1, now renamed
GFP signals were located in the nucleus and mito-
Deg1, has been shown to be proteolytically active
chondria, respectively (Supplemental Fig. S1). Thus,
toward thylakoid lumen proteins such as plastocyanin
these results indicate that Deg7 is targeted to the
and PsbO of PSII in vitro (Chassin et al., 2002). A 5.2-
kD C-terminal fragment of the D1 protein was de-
Next, intact chloroplasts were isolated, separated
tected in vitro after incubation of recombinant Deg1
into chloroplast stroma and thylakoid membrane frac-
with inside-out thylakoid membranes. In transgenic
tions, and then subjected to immunoblot analysis with
plants with reduced levels of Deg1, fewer of its 16-
a specific antibody against Deg7. The purity of chlo-
and 5.2-kD degradation products were observed
roplasts was confirmed by immunoblot analysis using
(Kapri-Pardes et al., 2007). Deg5 and Deg8 form a
the antibodies against the nuclear histone H1 protein,
dodecameric complex in the thylakoid lumen, and
the mitochondrial NDUFS1 protein, the cytosolic
recombinant Deg8 is able to degrade the photodam-
DHAR2 protein, and the thylakoid membrane protein
aged D1 protein of PSII in an in vitro assay (Sun et al.,
LHCII (Supplemental Fig. S2). The identities of the
2007a). The 16-kD N-terminal degradation fragment of
different fractions were further verified by immuno-
the D1 protein was detected in wild-type plants but
blot analyses with the antibodies against the thylakoid
not in a deg5 deg8 double mutant after high-light
membrane protein LHCII, the peripheral protein
treatment. The deg5 deg8 double mutant showed in-
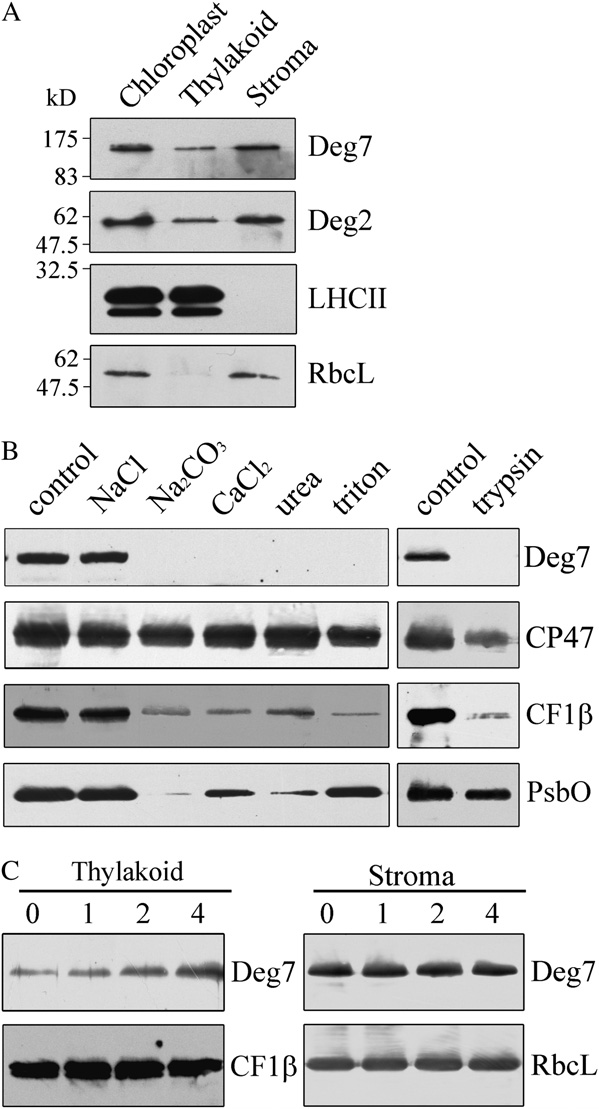
Deg2, and the stromal protein RbcL (Fig. 1A). To
creased sensitivity to high light and high temperature
determine the localization of Deg7, polyclonal antise-
in terms of growth and PSII activity compared with the
rum was raised and immunoblot analysis of chloro-
single mutants deg5 and deg8, suggesting that Deg5
plast protein showed the specificity of the Deg7
and Deg8 have overlapping functions in the primary
antibody (Supplemental Fig. S3). We found that Deg7
cleavage of the CD loop of the D1 protein (Sun et al.,
is located in chloroplasts and that the majority of
2007a, 2007b). In vitro analysis has demonstrated that
Deg7 is present in chloroplast stromal fractions (Fig.
recombinant stroma-localized Deg2 was also shown to
1A). To further investigate whether Deg7 is a periph-
be involved in the primary cleavage of the DE loop of
eral or an intrinsic membrane protein, the thylakoid
the D1 protein (Haußu¨hl et al., 2001). However, anal-
membranes were incubated with trypsin. Protease
ysis of a mutant lacking Deg2 suggested that Deg2
digestion assays showed that Deg7 could be degraded
Plant Physiol. Vol. 152, 2010
PSII Protein Degradation by DEG7
protein and is associated with the membranes. Next, toexamine the strength of Deg7's membrane association,we treated the thylakoid membranes with salts andchaotropic agents. Washing the membranes with 0.25M NaCl did not release the Deg7 from the membranes,but Deg7 was barely detectable after washing themembranes with 0.2 M Na CO , 1
As a control, we found that the integral membraneprotein CP47 was not released from the membranes bysuch treatments. High-light treatment increased theamount of Deg7 associated with the thylakoid mem-branes, and the levels of Deg7 in the stromal fractionsremained almost unchanged after exposure to high-light treatment (Fig. 1C). This may reflect an increasedexpression of Deg7 after high-light treatment.
Proteolytic Activity and Physiological Targets of Deg7
The increased association of Deg7 with the mem-
branes suggested that it might function in the degra-dation of photosynthetic proteins. To examine theproteolytic activity of the recombinant His-taggedDeg7, we incubated it with a mixture of a-, b-, and
k-forms of casein. b-Casein is the preferred substratefor assaying bacterial DegP activity under in vitroconditions (Lipinska et al., 1990). b-Casein was effi-ciently degraded by the fractions containing overex-pressed Deg7, and the a- and k-forms remainedunchanged under our experimental conditions (Sup-plemental Fig. S4). More than 50% of the b-casein wasdegraded within the first 30 min. These results indi-cated that the recombinant Deg7 was proteolyticallyactive.
Next, we examined whether Deg7 is proteolytically
active toward photosynthetic proteins. Since the DegPprotease has been shown to degrade damaged ormisfolded proteins (Gottesman, 1996; Pallen andWren, 1997; Clausen et al., 2002), the thylakoid mem-
Figure 1. Immunolocalization and expression of Deg7. A, Intact
branes used in this experiment were first subjected to
chloroplasts were separated into stroma and thylakoid membranes.
high-light treatment (1,800 mmol m22 s21) for 90 min at
Proteins in each fraction were resolved by SDS-PAGE, and the location
0°C. Treatment with high light causes irreversible
of Deg7 was determined by immunoblot analysis. B, The thylakoid
oxidative protein damage, which subsequently in-
membranes from mature Arabidopsis leaves were treated with trypsin
duces conformational changes in proteins, making
for 30 min at 25°C or incubated with 250 mM NaCl, 200 mM Na2CO3,
them vulnerable to proteolytic degradation (Prasil
M urea, and 0.05% Triton X-100 for 30 min at 4°C. After
these treatments, the thylakoid proteins were separated by SDS-PAGE
et al., 1992; Aro et al., 1993; Andersson and Aro,
and immunodetected with anti-Deg7, anti-CF1b, anti-PsbO, and anti-
2001). The thylakoid membranes isolated from leaves
CP47 antibodies. CF1b, PsbO, and CP47 were used as markers.
exposed to high light were treated with 50 mmol
Membrane preparations that had not been subjected to any of these
EDTA to inhibit the endogenous FtsH proteases. They
treatments were used as controls. The experiments were repeated five
were then incubated with or without recombinant
times independently, and similar results were obtained. Results of a
Deg7 in the dark at 37°C. In the absence of recombi-
representative experiment are shown. C, Expression of Deg7 associated
nant Deg7, the D1, D2, CP43, and CP47 protein levels
with the thylakoid membranes. The detached leaves, floating adaxial
remained unchanged and no degradation products
side up on water, were illuminated with a photon flux density of 1,800
were detected (Fig. 2A). However, when recombinant
mmol m22 s21 for 1, 2, and 4 h. Then, the thylakoid and stroma proteins
Deg7 was added to isolated thylakoid membranes, the
were prepared, and the proteins were subjected to SDS-PAGE andimmunoblot analyses.
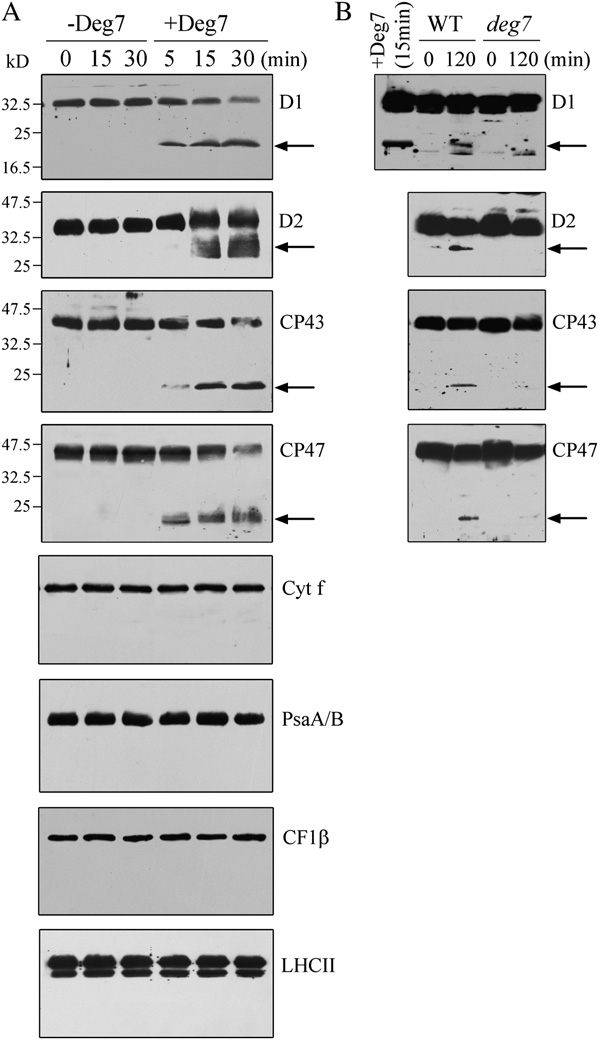
amounts of full-length D1, D2, CP43, and CP47 pro-teins decreased with time, and polypeptides of 20 kDfor D1, 29 kD for D2, 19 kD for CP43, and 19 kD for
by trypsin, while the lumen protein PsbO was pro-
CP47 were immunodetected using antibodies raised
tected from trypsin treatment (Fig. 1B). These results
against the DE loop of the D1 protein, D2 protein,
indicate that the Deg7 protein is a chloroplast stroma
CP43 protein, and CP47 protein, respectively (Fig. 2A).
Plant Physiol. Vol. 152, 2010
using isolated PSII and anti-CP47 for calibration (datanot shown), which showed a large excess of PSII overprotease.
Functions of PDZ Domains with Respect to the Activitiesof Deg7
To determine the function of the PDZ domains in
regulating the proteolytic activities of Deg7, we ex-pressed a series of Deg7 proteases that contain zero totwo PDZ domains in vitro (Supplemental Figs. S6–S8).
Regardless of the presence of either one or two PDZdomains in the truncated Deg7 protein, the levels ofPSII core proteins D1, D2, CP43, and CP47 decreasedand the specific degradation fragments of their corre-sponding proteins were not detected. When the trun-cated Deg7 without any PDZ domains was incubatedwith the thylakoid membranes, the amounts of notonly PSII core proteins D1, D2, CP43, and CP47 butalso LHCII, PsaA/B, Cyt f, and CF1b were reduced.
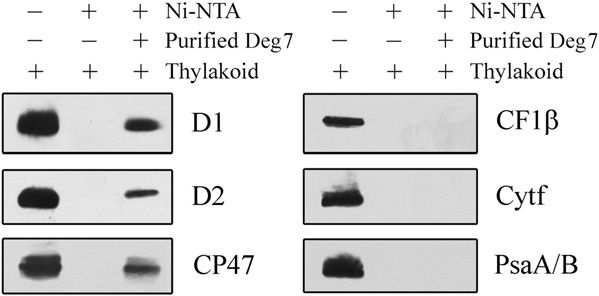
Association of Deg7 with PSII
The proteolytic activity of Deg7 toward PSII core
proteins suggests that Deg7 is associated with PSII. Totest this possibility, the thylakoid protein complexesseparated on a blue native gel were further subjectedto denaturing SDS-PAGE and immunoblot analysisusing specific antibodies. Our results showed that theDeg7 protein and PSII complex comigrated on the gel(Supplemental Fig. S9). Next, pull-down experimentswere performed with recombinant Deg7 fused to anN-terminal His tag. The purified His-Deg7 fusionprotein was incubated with n-dodecyl b-D-maltoside(DM)-solubilized thylakoid membranes. After wash-
Figure 2. Degradation of photodamaged PSII proteins by recombinant
ing nickel-nitrilotriacetic acid agarose (Ni-NTA) resin
Deg7. A, High-light-treated thylakoid membranes (10 mg of chloro-
with buffer, the bound proteins were separated by
phyll) were incubated with (+Deg7) or without (2Deg7) purified
SDS-PAGE and examined by immunoblot analysis
recombinant Deg7 (0.5 mg), and the degradation of thylakoid proteins
(Fig. 3). The PSII proteins D1, D2, and CP47 were
was detected by SDS-PAGE and immunoblot analyses with anti-D1,
detected when the His-Deg7 fusion protein was used
anti-D2, anti-CP43, anti-CP47, anti-Cyt f, anti-CF1b, anti-PsaA/B, andanti-LHCII antibodies. Molecular mass markers (kD) are to the left. B,Thylakoid membranes isolated from wild-type (WT) and deg7 leaveswere exposed to high-light illumination for 2 h. The thylakoid proteinswere separated by SDS-PAGE and immunodetected with antibodiesraised against the D1, D2, CP43, and CP47 proteins. The degradationfragments are indicated with arrows. Similar results were obtained intwo additional independent experiments. Results from a representativeexperiment are shown.
Immunoblot analysis with anti-Cyt f, anti-LHCII, anti-CF1b, and anti-PsaA/B antibodies showed that thelevels of these proteins remained almost constant in
the presence of recombinant Deg7 (Fig. 2A). When the
Interaction of Deg7 with PSII. Ten micrograms of the Deg7-
His tag fusion protein coupled to Ni-NTA resin was incubated with 100
thylakoid membranes were isolated from the plants
mg of DM-solubilized thylakoid membranes. Bound proteins were
that were not exposed to high-light treatment, the
eluted, separated by SDS-PAGE, and subjected to immunoblot analysis
photosynthetic proteins were not degraded by recom-
with anti-PsaA/B, anti-D1, anti-D2, anti-CP47, anti-Cyt f, and anti-
binant Deg7 (Supplemental Fig. S5). The relative mole
CF1b antibodies. Similar results were obtained in two additional
ratio of Deg7 and PSII was estimated to be about 1:10
Plant Physiol. Vol. 152, 2010

PSII Protein Degradation by DEG7
in the assay. In contrast, the PSI protein PsaA/B, the
Increased Sensitivity of the deg7 Mutants to High Light
protein Cyt f, and the ATP synthase protein
CF1b were not detected when the solubilized thyla-
To determine whether Deg7 is involved in photo-
koid membrane and the resin were incubated in the
inhibition and repair processes, the F /F
presence or absence of His-Deg7 (Fig. 3).
sured in the wild-type and deg7 plants under high-lightillumination (1,800 mmol m22 s21). In the absence oflincomycin, within 2 h of illumination at a lightintensity of 1,800 mmol m22 s21, F /F declined in the
T-DNA Insertion Mutants of deg7
wild-type and mutant leaves to about 53% and 39%,
To study the in vivo function of Deg7, we obtained
respectively, of the dark-adapted values (Fig. 5A).
Arabidopsis lines from the SALK collection. The deg7
These results clearly demonstrate the increased pho-
mutant line (SALK_075584) contains a T-DNA inser-
tosensitivity of the mutants. In the presence of linco-
tion within the 1,417-bp sequence downstream of the
mycin, the decline of F /F in the wild-type leaves was
ATG codon, and this was confirmed by PCR and
more rapid and continued until F /F
subsequent sequencing of the amplified products
proached about 10% of the dark-adapted values (Fig.
(Supplemental Fig. S10, A and B). Reverse transcrip-
5A). Interestingly, in the presence of lincomycin, the
tion (RT)-PCR analysis showed that expression of the
decline in F /F in the deg7 mutants was similar to that
Deg7 gene was not detectable (Supplemental Fig.
observed in the wild-type leaves during the same
S10C). Further immunoblot analyses revealed that
photoinhibitory light treatment (Fig. 5A). Since linco-
Deg7 protein was not detectable in the deg7 mutant
mycin blocks the repair of PSII by inhibiting de novo
and that the levels of Deg7 in the complemented plants
protein synthesis in the chloroplast, these results sug-
were comparable to those in the wild-type plants
gest that the wild-type and mutant leaves display
(Supplemental Fig. S10D). When grown under 120
similar rates of PSII photoinhibition.
mmol m22 s21 light, the growth rates of the deg7
To establish whether the high susceptibility of the
mutants were comparable to those of the wild type
deg7 mutants to photoinhibition is related to PSII
(Fig. 4A). Chlorophyll fluorescence analysis showed
protein turnover, we analyzed the PSII protein con-
that the maximal photochemical efficiency of PSII
tents of the thylakoid membranes in the wild-type and
(F /F ) was similar between the deg7 plants (0.82 6
mutant plants under high-light illumination in the
0.02) and wild-type plants (0.83 6 0.01).
presence of lincomycin. Under high-light illumination
To examine the steady-state levels of thylakoid
and in the presence of this inhibitor, the levels of the
proteins, immunoblot analyses were performed with
PSII proteins D1, D2, CP43, and CP47 gradually de-
antibodies raised against specific subunits of the pho-
clined in the deg7 and wild-type plants, but the rate of
tosynthetic protein complexes (Supplemental Fig.
reduction was slower in the mutant (Fig. 5B). The
S11). Our results showed that levels of the thylakoid
levels of PsbO, another PSII protein, and the PSI
proteins, including D1, D2, LHCII, PsbO, and CP43 of
protein PsaA/B were found to be relatively stable in
PSII, PsaA/B of PSI, Cyt f of the Cyt b f complex, and
both the wild-type and mutant plants.
CF1b of ATP synthase, were not altered in the deg7
Next, we performed an in vivo labeling experiment
mutant. The levels of Deg1, Deg5, Deg8, and FtsH
to follow the turnover of newly synthesized chloro-
were also not changed in the mutant.
plast proteins (Fig. 5C). Our results showed that the
Figure 4. Phenotypes of deg5, deg7, deg8, deg2 deg7, deg5 deg7, and deg8 deg7 mutants and wild-type (WT) plants. A, Four-week-old deg5, deg7, deg8, deg2 deg7, deg5 deg7, deg8 deg7, and deg7 complemented mutants and wild-type plants grownunder a photon flux density of 120 mmol m22 s21. B, Two-week-old deg5, deg7, deg8, deg2 deg7, deg5 deg7, deg8 deg7, anddeg7 complemented mutants and wild-type plants grown in a growth chamber under a photon flux density of 120 mmol m22 s21and then transferred to a greenhouse for another 2 weeks with a maximum photon flux density at noon of about 1,000 mmolm22 s21.
Plant Physiol. Vol. 152, 2010
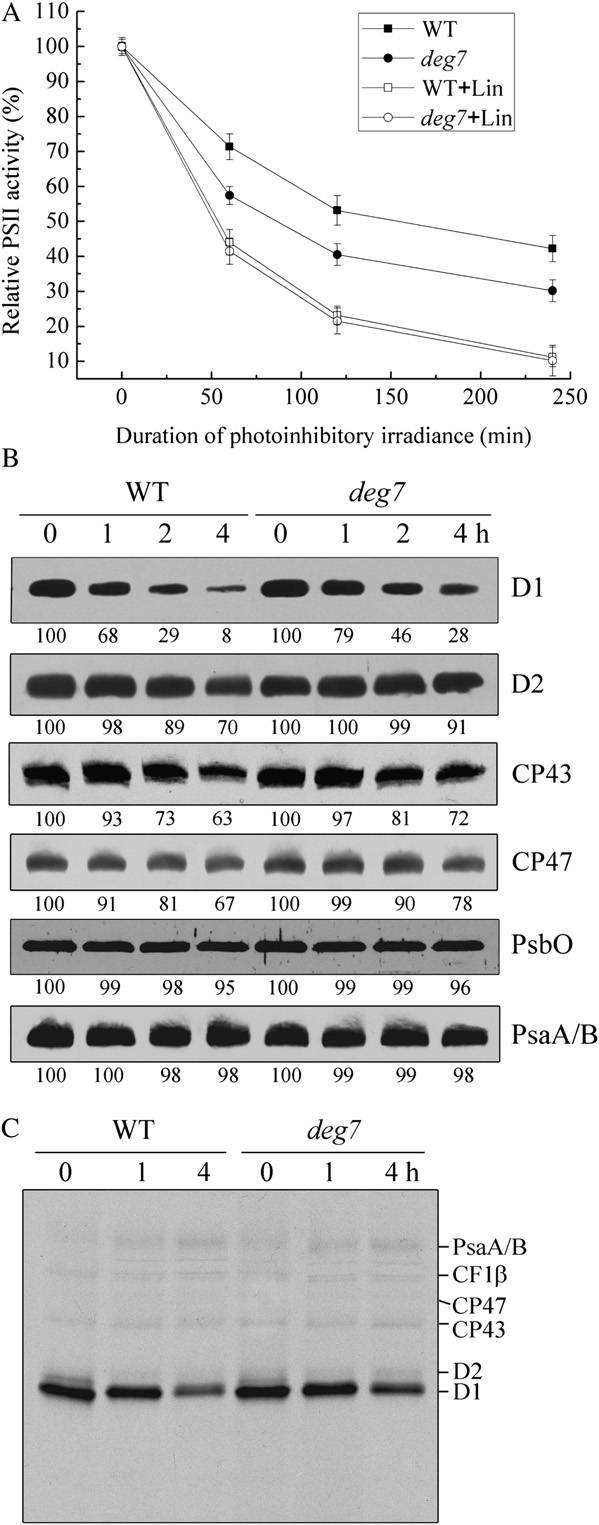
synthesis rates of the PSII core proteins D1, D2, CP47,and CP43, the PSI reaction center proteins PsaA/B,and the ATP synthase CF1a/b were similar betweenthe wild-type and mutant plants. The PSII core pro-teins D1, D2, CP47, and CP43 showed increased sta-bility in the mutant compared with the wild-typeplants. The protein synthesis rates in pulse labeling for20 min reflect the differences between the synthesisand degradation rates of photosynthetic proteins.
Thus, the net synthesis rate of PSII reaction centerprotein D1 in the mutant was lower than that of thewild type, since the rate of D1 degradation was slowedin deg7.
To further analyze the physiological function of
Deg7, photoinhibition was carried out with isolatedthylakoid membranes to which 50 mmol EDTA hadbeen added in order to inactivate the endogenous FtsHproteases. We found that the 20-kD polypeptide of D1,the 29-kD polypeptide of D2, the 19-kD polypeptide ofCP43, and the 19-kD polypeptide of CP47 were de-tected in the samples from wild-type plants follow-ing photoinhibition but not in samples from the deg7mutant. An 18-kD polypeptide from D1 was detectedboth in the wild-type and mutant plants (Fig. 2B),which may be due to the function of lumen-localizedDeg5 and Deg8 (Sun et al., 2007a).
Since Deg7 is involved in protection against photo-
inhibition, we analyzed the phenotypes of the mutantsduring growth under high irradiance. For this pur-pose, we transferred the wild-type and deg7 mutantplants that were grown initially at 120 mmol m22 s21 toa greenhouse with a maximum intensity of 1,000 mmolm22 s21 at noon. Our results showed that when ex-posed to high light, the growth of the deg7 mutantwas inhibited compared with the wild-type plants(Fig. 4B).
Generation of deg2 deg7, deg5 deg7, and deg8 deg7 DoubleMutants and Their Growth under High Light
Three double mutants were constructed, deg2 deg7,
deg5 deg7, and deg8 deg7, to test the physiologicalimportance of stromal and luminal Deg proteases inphotosynthesis. The growth rates and PSII activities ofdeg2 deg7, deg5 deg7, and deg8 deg7 were comparableto those of wild-type plants when grown at 120 mmol
tected with specific antibodies directed against D1, D2, CP43, CP47,PsbO, and PsaA/B proteins. X-ray films were scanned and analyzedusing an AlphaImager 2200 documentation and analysis system. The
Figure 5. Changes in PSII activity and protein levels. A, The Fv/Fm was
percentage protein levels shown below the lanes were estimated by
measured for detached leaves from wild-type (WT) plants (squares) and
comparison with levels found in corresponding samples taken at time
deg7 mutant plants (circles) following high-light illumination (1,800
0. Similar results were obtained in two additional independent exper-
mmol m22 s21) in the absence (white symbols) or presence (black
iments. C, Pulse-chase analysis of chloroplast proteins. Two-week-old
symbols) of lincomycin (Lin). B, Immunoblot analysis of thylakoid
leaves of the wild-type and deg7 mutant plants were labeled for 20 min
proteins following high-light illumination. Thylakoid membranes were
followed by a chase after 1 or 4 h at 120 mmol m22 s21. Thylakoid
isolated from the wild-type and deg7 mutant leaves during exposure to
membranes with equal amounts of chlorophyll were isolated and
high-light illumination in the presence of lincomycin for 1, 2, or 4 h.
separated by SDS-PAGE. The resolved proteins were visualized by
The thylakoid proteins were separated by SDS-PAGE and immunode-
Plant Physiol. Vol. 152, 2010
PSII Protein Degradation by DEG7
m22 s21 (Fig. 4A). However, when exposed to high
5B). Pulse-chase labeling experiments also revealed
light, the growth of the mutants was slower than that
that the turnover rates of newly synthesized PSII core
of the wild-type plants, and the extent of the pheno-
proteins were slowed in the deg7 mutant (Fig. 5C).
type in the deg5 deg7 and deg8 deg7 double mutants was
These results suggest that Deg7 is involved in the
more pronounced than in the single mutants, while
degradation of PSII core proteins.
that of deg2 deg7 was similar to that of the deg7 mutant
Deciphering the physiological roles of specific pro-
(Fig. 4B). Moreover, some of the mutant leaves ex-
teases is intimately related to the identification of their
hibited symptoms of yellowing in the deg5 deg7 and
substrates. The D1 protein of the PSII reaction center
deg8 deg7 double mutants (Fig. 4B).
has been identified as the substrate of characterizedchloroplast Deg proteases in vivo. This protein issusceptible to proteolytic cleavage at its lumen-exposed domains (the AB and CD loops and the C
terminus) by Deg1 (Kapri-Pardes et al., 2007) and theDeg5-Deg8 complex (Sun et al., 2007a). The detection
The adverse effect of high light on plant growth in
of a C-terminal D1 fragment of about 20 kD in in vitro
the deg7 mutant indicates the physiological impor-
experiments suggests that Deg7 catalyzes the cleavage
tance of Deg7 for photoprotection under high-light
of the photodamaged D1 protein at the stromal loop
illumination (Figs. 4 and 5). Such a role for Deg7 is also
connecting the B and C transmembrane helices (Fig. 2).
in agreement with our observation of a dramatic
Another stroma-localized Deg2 protein was shown to
increase in the levels of proteins associated with the
be involved in the initial cleavage of the DE loop of the
thylakoid membranes under high light based on im-
D1 protein in vitro (Haußu¨hl et al., 2001); however, this
munoblot analysis (Fig. 1). There was no apparent
possibility is not supported in vivo, since the deg2
difference in growth between wild-type and mutant
mutant showed a similar extent of PSII inactivation
plants under normal growth conditions at 120 mmol
and a similar D1 protein turnover rate to that in the
m22 s21, which also reflected the similar F /F ratios.
wild-type plants (Huesgen et al., 2006). Since Deg2 and
When the wild-type plants were subjected to high-
Deg7 are located in chloroplast stroma, we analyzed a
light treatment, the PSII activity was reduced for the
deg2 deg7 double mutant, and the deg2 deg7 and deg7
duration of treatment in the absence of lincomycin,
mutants showed similar degrees of sensitivity to high
and the reduction of PSII activity became more pro-
light in terms of growth (Fig. 4) and PSII activity
nounced in the presence of lincomycin (Fig. 5). The
(Supplemental Fig. S12). Thus, Deg2, unlike Deg7,
PSII activity in the deg7 mutant showed enhanced
seems to have little effect on PSII repair in vivo.
sensitivity to high-light treatment in the absence of
Although the PSII reaction center protein D1 is best
lincomycin compared with the wild-type plants, while
characterized as the main target of photodamage, the
the rate of PSII photoinhibition was similar in the
other PSII proteins D2, CP47, and CP43 are occasion-
mutant and wild-type plants in the presence of the
ally damaged and degraded, especially in response
protein synthesis inhibitor lincomycin (Fig. 5). These
to increasing light intensity (Schuster et al., 1988;
results indicate that under high light, the repair of PSII
Yamamoto and Akasaka, 1995; Jansen et al., 1999;
was perturbed in the mutant. It is likely that the
Adir et al., 2003). The detection of the 29-kD fragment
increased sensitivity of PSII to high light in the mutant
of D2 indicates that the cleavage site of photodamaged
could be ascribed to the impairment of the PSII repair
D2 is located at its N terminus. The observation of the
19-kD fragments of CP47 and CP43 after treatment of
The repair of PSII involves the degradation and
thylakoid membranes with recombinant Deg7 sug-
removal of damaged PSII proteins and their subse-
gests that cleavage may take place at the stromal loop
quent replacement with newly synthesized copies
connecting transmembrane domains D and E.
(Prasil et al., 1992; Aro et al., 1993; Andersson and
The PDZ domains in E. coli DegP proteases have
Aro, 2001). The degradation of photodamaged PSII
been implicated in the formation of oligomers and also
proteins is inherent to the repair cycle of PSII after
in the regulation of proteolytic activity (Doyle et al.,
photoinhibition. Removal of photooxidatively dam-
1996; Harris and Lim, 2001). It is interesting that Deg7
aged PSII proteins and subsequent replacement with
harbors three PDZ domains. The presence of three
newly synthesized copies represents an efficient repair
PDZ domains raises questions about their functions.
cycle. However, the detailed mechanisms of the repair
Truncated Deg7 containing either one or two PDZ
of PSII are far from being understood. Impairment of
domains was also able to degrade PSII core proteins,
the PSII repair cycle could result from the perturbation
but it did not generate specific fragments. The trun-
of either protein synthesis or degradation. Our pulse-
cated Deg7 that lacked any PDZ domain lost its
labeling experiments showed that the synthesis rates
specificity for the substrate and was proteolytically
of the PSII core proteins D1, D2, CP47, and CP43 were
active not only toward PSII core proteins but also
not affected in the mutant (Fig. 5C). Instead, exami-
toward other photosynthetic proteins (Supplemental
nation of the degradation of PSII core proteins showed
Figs. S6–S8). These results highlight the importance of
that the decrease in the levels of PSII core proteins was
PDZ domains as regulators of proteolytic activity that
slower in the mutant than in the wild-type plants (Fig.
recognize and discriminate between substrates.
Plant Physiol. Vol. 152, 2010
The function of Deg7 in the degradation of photo-
the D1 protein (Komenda et al., 2006). These studies
damaged PSII core proteins suggests its possible in-
suggest a possible function of FtsH in the degradation
teraction with PSII. The results presented in Figure 3
of other PSII core proteins. Considering the functional
and Supplemental Figure S9 strongly suggest that
mode of FtsH, it is likely that other PSII core proteins
Deg7 associates with PSII complexes but not with
may also be cleaved by peptidases in order to gener-
other photosynthetic complexes such as PSI, Cyt b f, or
ate limited numbers of distinct fragments, followed
ATP synthase. This association is consistent with the
by their complete digestion by the processive ATP-
physiological role of Deg7 in the repair of damage to
dependent FtsH protease.
PSII caused by photoinhibition. The finding of anassociation between a protease and a complex con-taining its substrates is intriguing. Our results suggestthat, at least in the case of PSII (which is known to
MATERIALS AND METHODS
undergo continuous cycles of assembly and repair),
protein complexes might be equipped with compo-nents that are essential for their repair.
Wild-type and mutant Arabidopsis (Arabidopsis thaliana ecotype Columbia)
The distribution of Deg proteases on both the stro-
plants were grown in soil under short-day conditions (10-h-light/14-h-darkcycles) with a photon flux density of 120 mmol m22 s21 at a constant
mal and luminal sides of the thylakoid membranes
temperature of 22°C. The T-DNA insertion line SALK_075584 (Deg7;
might have implications for the efficient degradation
At3g03380) was obtained from the T-DNA-transformed Arabidopsis collection
of damaged D1 protein. It is thus reasonable to spec-
from the Arabidopsis Biological Resource Center (Ohio State University).
ulate that Deg7 functions cooperatively with lumen-
Homozygous deg7 mutant plants were identified by PCR analyses using thefollowing gene-specific and T-DNA-specific primers: LP (5#-GGTACGTT-
localized Deg1, Deg5, and Deg8, generating more
CAGTGACACACCC-3#), RP (5#-CAAAGCTTTCTAGGGGTGCTC-3#), and
cleavage sites on both sides of the membranes. This
T-DNA LB (5#-GCGTGGACCGCTTGCTGCAACT-3#). The precise location of
may increase the efficiency of D1 degradation. In mito-
the T-DNA insertion was determined by sequencing the PCR products.
chondria, the degradation of membrane proteins was
Confirmation of null mutants was carried out by RT-PCR using the same
greatly assisted by the cooperation of proteases func-
primer set described above for Deg7. Equal cDNA loading in each sample wasmonitored by RT-PCR analysis of the expression level of actin (5#-AACTGG-
tioning on both sides of the membranes (Leonhard
GATGATATGGAGAA-3# and 5#-CCTCCAATCCAGACACTGTA-3#). The
et al., 2000). The single mutants deg5, deg7, and deg8
double mutants deg2 deg7, deg5 deg7, and deg7 deg8 were obtained by PCR
all showed decreased growth compared with the wild-
screening of an F2 population from crossed single mutant lines.
type plants under high light, although these mutantsdid not show any apparent differences in terms
Complementation of the deg7 mutant
of phenotype under normal growth conditions at
The cDNA containing the coding region of Deg7 was amplified with the
120 mmol m22 s21 (Fig. 4; Sun et al., 2007a), which
following primers, which include XhoI and KpnI restriction sites at their
indicates that the Deg proteases are important for
5# ends to facilitate cloning: sense primer (5#-GACCCGGGGGAGAT-
efficient PSII repair. The additive functions of Deg
CAAATGGGAGATCC-3#) and antisense primer (5#-GGGGTACCTTACTG-
proteases were also demonstrated by the phenotypes
CAAGGCTTTCAATATA-3#). The resulting fragment was cloned into the SamI
of the deg5 deg7 and deg8 deg7 double mutants grown
and KpnI sites of pSN1301 under the control of the cauliflower mosaic virus35S promoter. The plasmid pSN1301-Deg7 was transformed into Agrobacte-
under high light.
rium tumefaciens strain C58 via electroporation and introduced into the
Since Deg proteases are endopeptidases that can
homozygous deg7 plants (Clough and Bent, 1998). Transformant plants were
cleave intermembrane peptide regions exposed to the
selected on medium containing half-strength Murashige and Skoog salt mix,
stromal or the luminal side, it is unlikely that PSII
50 mg mL21 hygromycin, and 0.8% agar. The resistant plants were transferredto soil to grow to maturity, and their transgenic status was further confirmed
proteins are completely degraded by Deg proteases.
by PCR and immunoblot analyses.
The creation of D1 degradation intermediates mayprovide additional sites for the initiation of proteoly-
sis. The function of FtsH in degrading E. coli integralmembrane proteins involves relocation of regions of
Detached mature leaves were floated adaxial side up on water and
substrates from one side to the other (Ito and Akiyama,
exposed to a photon flux density of 1,800 mmol m22 s21, and chlorophyll
2005). It may be that Deg proteases from both sides of
fluorescence was measured using a PAM-2000 fluorometer (Walz). Thetemperature of the water was kept at 22°C during photoinhibition treatments.
the membranes enhance the efficiency of D1 degrada-
Synthesis of chloroplast-encoded proteins was blocked by incubating de-
tion by increasing the number of D1 degradation
tached leaves with their petioles submersed in 1 mM lincomycin solution at an
intermediates that are accessible by FtsH. The Arabi-
irradiance of 20 mmol m22 s21 for 3 h prior to photoinhibitory light treatment.
dopsis var2 mutant lacking FtsH2 exhibited more
To investigate the effects of high irradiance on plant growth, we transferred
2-week-old Arabidopsis plants grown in a growth chamber under a photon
increased sensitivity to high-light treatment than the
flux density of 120 mmol m22 s21 to a greenhouse under sunlight illumination
deg7 mutant (Supplemental Fig. S12), which suggests
for another 2 weeks. In the greenhouse, the maximum photon flux density at
that FtsH2 is predominantly involved in the PSII repair
noon was about 1,000 mmol m22 s21 and average day/night temperatures
cycle. The light-dependent degradation of D1 and D2
were 25°C/22°C.
was slowed in the Arabidopsis var2 mutant (Baileyet al., 2002). In Synechocystis species PCC6803, the FtsH
Thylakoid Membrane Preparation
protease slr0228, which shows the highest similarity to
Thylakoid membranes were isolated essentially as described by Zhang
Arabidopsis FtsH2, has been shown to play a more
et al. (1999). Briefly, Arabidopsis leaves were homogenized by a mortar and
general role in the removal of PSII proteins other than
pestle in an ice-cold isolation buffer (400 mM Suc, 50 mM HEPES-KOH, pH 7.8,
Plant Physiol. Vol. 152, 2010
PSII Protein Degradation by DEG7
10 mM NaCl, and 2 mM MgCl ), filtered through two layers of cheesecloth, and
Proteolytic Degradation Assays
centrifuged at 5,000g for 10 min. The thylakoid pellets were resuspended inthe isolation buffer and then centrifuged at 5,000g for 10 min. The thylakoids
The proteolytic activity was assayed by incubating Deg7 with 15-mg
were finally suspended in isolation buffer, and their chlorophyll levels were
mixtures of casein (a-, b-, and k-casein; Sigma-Aldrich) in standard reaction
mixtures including 0.5 mg of purified Deg7 in a 30-mL solution containing 50mM Tris-HCl, pH 7.6. The mixtures were incubated for 0, 15, 30, 45, and 60 minat 37°C, separated by SDS-PAGE, and stained with Coomassie Brilliant Blue.
SDS-PAGE and Immunoblot Analysis
For the in vitro degradation assay, wild-type Arabidopsis thylakoid
membranes were illuminated at 1,800 mmol m22 s21 for 90 min at 4°C and
The protein sample was mixed with the same volume of 23 SDS sample
centrifuged at 15,000g for 15 min. The collected thylakoids were washed with
buffer (125 mM Tris-HCl, pH 6.8, 20% [w/v] glycerol, 4% SDS, 5% b-mercap-
1.0 M CaCl2 to reduce endogenous Deg7 activity before the addition of
toethanol, and 0.1% bromphenol blue) for 60 min and layered onto 15% SDS
recombinant Deg7 or its derivatives and resuspended in 300 mM sorbitol and
polyacrylamide gels containing 6 M urea (Laemmli, 1970). After electropho-
10 mM HEPES-KOH, pH 8.0. After incubation of purified recombinant Deg7
resis, gels were stained with Coomassie Brilliant Blue. For immunoblot
protein with the thylakoid membranes at 37°C in the dark for various amounts
analysis, the proteins resolved by SDS-PAGE were blotted onto nitrocellulose
of time, the samples were subjected to SDS-PAGE and immunoblot analyses.
membranes and reacted with specific antibodies, and the signals werevisualized with the enhanced chemiluminescence method.
Primary antibodies used in this study were as follows: (1) a D1-specific
In Vivo Labeling of Chloroplast Proteins
antibody against the oligopeptide NYGYKFGQE containing the amino acids234 to 242 in the DE loop of the D1 protein of Synechocystis species PCC 6803;
In vivo chloroplast protein labeling was carried out essentially according to
(2) a D2-specific antibody against the oligopeptide NTFRAFNPTQAEETYS
Meurer et al. (1998). Leaves of 2-week-old Arabidopsis plants were preincu-
containing the amino acids 231 to 246 in the DE loop of the D2 protein of
bated for 30 min in the presence of 20 mg mL21 cycloheximide, which blocks
Arabidopsis; (3) an anti-peptide antibody specific for E. coli FtsH, which is
the synthesis of nucleus-encoded proteins. Then, the leaves were radiolabeled
potentially cross-reactive with all Synechocystis species PCC 6803 FtsH homo-
with 1 mCi mL21 [35S]Met (specific activity . 1,000 Ci mmol21; Amersham
logs; (4) a Cyt f antibody raised against the amino acids 146 to 257 in the Cyt f
Pharmacia Biotech) at 120 mmol m22 s21 in the presence of 20 mg mL21
protein of Arabidopsis; (5) a CF1b antibody raised against the amino acids 206
cycloheximide for 20 min at 22°C, followed by a chase of 1 or 4 h in buffer
to 387 in the CF1b protein of Arabidopsis; (6) a PsaA/B antiserum raised
containing 10 mM cold Met. Afterward, the leaves were collected, the thyla-
against the amino acids 3 to 77 in the N terminus of the PsaA protein of
koid membranes were isolated, and the proteins were separated by SDS-
Arabidopsis; (7) a CP43 antibody raised against the amino acids 330 to 449 in
PAGE. For autoradiography, gels were stained, dried, and exposed to x-ray
the C terminus of the CP43 protein of Arabidopsis; (8) a CP47 antibody raised
against the amino acids 330 to 475 in the C terminus of the CP47 protein ofArabidopsis; (9) a LHCII antibody raised against the amino acids 25 to 154 inthe LHCB3 protein of Arabidopsis; (10) a Deg1 antibody raised against the
GFP Fusion Constructs for Transient Expression
amino acids 315 to 436 in the C terminus of the Deg1 protein of Arabidopsis;
(11) a Deg5 antibody raised against the amino acids 91 to 211 in the N terminusof the Deg5 protein of Arabidopsis; and (12) a Deg8 antibody raised against
A fragment encoding the N-terminal amino acids 1 to 243 of Deg7
the amino acids 297 to 423 in the C terminus of the Deg8 protein of
was amplified by PCR using the following primers: 5#-GCGTCGACAT-
GGGAGATCCGTTGGAGAG-3# and 5#-GGCCATGGTTAACGCCCTAACA-ACTCG-3#. The PCR product was cloned into the SalI and NcoI sites ofexpression vector pUC18-35S-sGFP to generate a fusion protein with the GFP
Antiserum Production and Immunolocalization Studies
as a reporter in the C terminus. The transit peptide (amino acids 1–245) of theFtsH11, the N-terminal part (amino acids 1–282) of AtFbr1, and the entire
The His-Deg7 fusion protein was purified on a Ni-NTA resin matrix, and
coding region of FRO1 were used as chloroplast, nuclear, and mitochondrial
polyclonal antibodies were raised in rabbit with the purified antigens. The
controls, respectively (Sakamoto et al., 2003; Cai et al., 2009). Isolation of the
intracellular localization of Deg7 was determined essentially according to
Arabidopsis protoplast and polyethylene glycol-mediated transfection were
Lennartz et al. (2001). The Arabidopsis membranes were suspended to a final
performed as described by Sakamoto et al. (2003). The cells with GFP signals
concentration of 50 mg chlorophyll mL21 in 10 mM HEPES-KOH, pH 8.0, 10
were examined using a confocal laser scanning microscope (LSM510; Carl
M sorbitol, and 1 mM phenylmethylsulfonyl fluoride
supplemented with 250 mM NaCl, 200 mM Na2CO3, 1 M CaCl2, 6 M urea, and0.05% Triton X-100 at 0°C for 30 min or 0.05 mg mL21 trypsin at 25°C for 30min. Membrane fractions without supplements were used as a control. After
Sequence data from this article can be found in the GenBank/EMBL data
treatment, the membranes were pelleted at 100,000g for 2 h at 4°C, quickly
libraries under accession number At3g03380 (DEG7).
washed with isolation buffer, and used for SDS-PAGE and immunoblotanalysis.
Supplemental Data
Recombinant Expression of Deg7 and Pull-Down Assays
The following materials are available in the online version of this article.
Full-length Deg7 cDNA was cloned into the pET28a plasmid and trans-
Supplemental Figure S1. Subcellular localization of the Deg7 protein.
formed into BL21 cells. The expression of the His-Deg7 fusion protein was
Supplemental Figure S2. Immunoblot analysis of the purity of isolated
induced by isopropylthio-b-D-galactoside (0.4 mM) for 2 h, and the overex-
pressed protein was purified using a Ni-NTA resin matrix. The purifiedprotein was renatured through a Sephadex G-75 column by eluting with 20
Supplemental Figure S3. Immunoblot analysis of chloroplast proteins
with the specific Deg7 antibody.
2PO4, pH 7.8, buffer, and the identity of the protein was confirmed by
immunoblot analyses with specific antibodies.
Supplemental Figure S4. Engineering of the Deg7 deletion construct and
For protein pull-down assays, thylakoid membranes (100 mg of chloro-
proteolytic activity of Deg7 with b-casein.
phyll) were solubilized with 1% (w/v) DM in 20% (w/v) glycerol, 25 mMBisTris-HCl, pH 7.0, and 1 mM phenylmethylsulfonyl fluoride for 15 min at
Supplemental Figure S5. Degradation of photosynthetic proteins by a
4°C, then centrifuged at 10,000g for 10 min. The supernatant obtained after
recombinant Deg7 protein.
centrifugation was incubated with His-Deg7 coupled to Ni-NTA resin. After
Supplemental Figure S6. Degradation of photodamaged PSII proteins by
incubation overnight with constant rotation at 4°C, the beads were washed
a recombinant Deg7 protein containing two PDZ domains.
five times with 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, and 1 mM EDTA buffer,and the bound proteins were eluted with SDS-PAGE sample buffer. The eluted
Supplemental Figure S7. Degradation of photodamaged PSII proteins by a
proteins were resolved by SDS-PAGE followed by immunoblot analyses.
recombinant Deg7 protein containing one PDZ domain.
Plant Physiol. Vol. 152, 2010
Supplemental Figure S8. Degradation of photodamaged PSII proteins by a
in cyanobacteria and chloroplasts of higher plants. Physiol Plant 123:
recombinant Deg7 protein lacking PDZ domains.
Huesgen PF, Schuhmann H, Adamska I (2006) Photodamaged D1 protein
Supplemental Figure S9. Immunoblot analysis of the association of Deg7
is degraded in Arabidopsis mutants lacking the Deg2 protease. FEBS Lett
with photosynthetic protein complexes after blue native/SDS-PAGE.
Supplemental Figure S10. Identification of the deg7 mutant.
Ito K, Akiyama Y (2005) Cellular functions, mechanism of action, and
regulation of FtsH protease. Annu Rev Microbiol 59: 211–231
Supplemental Figure S11. Analysis of thylakoid proteins from deg7
Itzhaki H, Naveh L, Lindahl M, Cook M, Adam Z (1998) Identification and
mutant and wild-type plants.
characterization of DegP, a serine protease associated with the luminal
Supplemental Figure S12. Changes in F /F
side of the thylakoid membrane. J Biol Chem 273: 7094–7098
m of wild-type, deg7, deg2 deg7,
and ftsh2 plants following high-light illumination.
Jansen MA, Mattoo AK, Edelman M (1999) D1-D2 protein degradation in
the chloroplast: complex light saturation kinetics. Eur J Biochem 260:527–532
Jiang J, Zhang X, Chen Y, Wu Y, Zhou ZH, Chang Z, Sui SF (2008)
Activation of DegP chaperone-protease via formation of large cage-likeoligomers upon binding to substrate proteins. Proc Natl Acad Sci USA
We thank Professor W. Sakamoto for helpful suggestions. We thank
105: 11939–11944
Professor Z. Adam for ftsh2 mutant seeds and anti-FtsH antibody. We are
Kapri-Pardes E, Naveh L, Adam Z (2007) The thylakoid lumen protease
grateful to the Arabidopsis Biological Resource Center for Arabidopsis seeds.
Deg1 is involved in the repair of photosystem II from photoinhibition inArabidopsis. Plant Cell 19: 1039–1047
Received November 9, 2009; accepted January 15, 2010; published January 20,
Kato Y, Sakamoto W (2009) Protein quality control in chloroplasts: a
current model of D1 protein degradation in the photosystem II repaircycle. J Biochem 146: 463–469
Kieselbach T, Funk C (2003) The family of Deg/HtrA proteases: from
Escherichia coli to Arabidopsis. Physiol Plant 119: 337–346
Kim DY, Kim DR, Ha SC, Lokanath NK, Lee CJ, Hwang HY, Kim KK
(2003) Crystal structure of the protease domain of a heat shock protein
Adam Z, Rudella A, van Wijk KJ (2006) Recent advances in the study of
HtrA from Thermotoga maritime. J Biol Chem 278: 6543–6551
Clp, FtsH and other proteases located in chloroplasts. Curr Opin Plant
Kim DY, Kim KK (2005) Structure and function of HtrA family proteins,
Biol 9: 234–240
the key players in protein quality control. J Biochem Mol Biol 38:
Adir N, Shochat S, Inoue Y, Ohad I (1990) Mechanism of the light-
dependent turnover of the D1 protein. J Biol Chem 265: 12563–12568
Komenda J, Barker M, Kuvikova´ S, de Vries R, Mullineaux CW, Tichy M,
Adir N, Zer H, Shochat S, Ohad I (2003) Photoinhibition: a historical
Nixon PJ (2006) The FtsH protease slr0228 is important for quality
perspective. Photosynth Res 76: 343–376
control of photosystem II in the thylakoid membrane of Synechocystis sp.
Andersson B, Aro EM (2001) Photodamage and D1 protein turnover in
PCC 6803. J Biol Chem 281: 1145–1151
photosystem II. In B Andersson, E-M Aro, eds, Regulation of Photosyn-
Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T (2002)
thesis. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp
Crystal structure of DegP (HtrA) reveals a new protease-chaperone
machine. Nature 416: 455–459
Aro EM, Virgin I, Andersson B (1993) Photoinhibition of photosystem II:
Krojer T, Sawa J, Scha¨fer E, Saibil HR, Ehrmann M, Clausen T (2008)
inactivation, protein damage and turnover. Biochim Biophys Acta 1143:
Structural basis for the regulated protease and chaperone function of
DegP. Nature 453: 885–890
Bailey S, Thompson E, Nixon PJ, Horton P, Mullineaux CW, Robinson C,
Kyle DJ, Ohad I, Arntzen CJ (1984) Membrane protein damage and repair:
Mann NH (2002) A critical role for the Var2 FtsH homologue of
selective loss of a quinone-protein function in chloroplast membranes.
Arabidopsis thaliana in the photosystem II repair cycle in vivo. J Biol
Proc Natl Acad Sci USA 81: 4070–4074
Chem 277: 2006–2011
Laemmli UK (1970) Cleavage of structural proteins during the assembly of
Bergantino E, Brunetta A, Touloupakis E, Segalla A, Szabo I, Giacometti
the head of bacteriophage T4. Nature 227: 680–685
GM (2003) Role of the PSII-H subunit in photoprotection: novel aspects
Lennartz K, Plu¨cken H, Seidler A, Westhoff P, Bechtold N, Meierhoff K
of D1 turnover in Synechocystis 6803. J Biol Chem 278: 41820–41829
(2001) HCF164 encodes a thioredoxin-like protein involved in the
Cai WH, Ji DL, Peng LW, Guo JK, Ma JF, Zou MJ, Lu CM, Zhang LX (2009)
biogenesis of the cytochrome b6f complex in Arabidopsis. Plant Cell 13:
LPA66 is required for editing psbF chloroplast transcripts in Arabidop-
sis. Plant Physiol 150: 1260–1271
Leonhard K, Guiard B, Pellecchia G, Tzagoloff A, Neupert W, Langer T
Chassin Y, Kapri-Pardes E, Sinvany G, Arad T, Adam Z (2002) Expression
(2000) Membrane protein degradation by AAA proteases in mitochon-
and characterization of the thylakoid lumen protease DegP1 from
dria: extraction of substrates from either membrane surface. Mol Cell 5:
Arabidopsis. Plant Physiol 130: 857–864
Clausen T, Southan C, Ehrmann M (2002) The HtrA family of proteases:
Li W, Srinivasula SM, Chai J, Li P, Wu JW, Zhang Z, Alnemri ES, Shi Y
implications for protein composition and cell fate. Mol Cell 10: 443–455
(2002) Structural insights into the pro-apoptotic function of mitochon-
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacte-
drial serine protease HtrA2/Omi. Nat Struct Biol 9: 436–441
rium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743
Lindahl M, Spetea C, Hundal T, Oppenheim AB, Adam Z, Andersson B
Doyle DA, Lee A, Lewis J, Kim E, Sheng M, Mackinno R (1996) Crystal
(2000) The thylakoid FtsH protease plays a role in the light induced
structures of a complexed and peptide-free membrane protein-binding
turnover of the photosystem II D1 protein. Plant Cell 12: 419–431
domain: molecular basis of peptide recognition by PDZ. Cell 85:
Lindahl M, Tabak S, Cseke L, Pichersky E, Andersson B, Adam Z (1996)
Identification, characterization, and molecular cloning of a homologue
Gottesman S (1996) Proteases and their targets in Escherichia coli. Annu Rev
of the bacterial FtsH protease in chloroplasts of higher plants. J Biol
Genet 30: 465–506
Chem 271: 29329–29334
Hagman A, Shi LX, Rintamaki E, Andersson B, Schroder WP (1997) The
Lipinska B, Sharma S, Georgopoulos C (1988) Sequence analysis and
nuclear-encoded PsbW protein subunit of photosystem II undergoes
regulation of the htrA gene of Escherichia coli: a sigma 32-independent
light-induced proteolysis. Biochemistry 36: 12666–12671
mechanism of heat-inducible transcription. Nucleic Acids Res 16:
Harris BZ, Lim WA (2001) Mechanism and role of PDZ domains in
signaling complex assembly. J Cell Sci 114: 3219–3231
Lipinska B, Zylicz M, Georgopoulos C (1990) The HtrA (DegP) protein,
Haußu¨hl K, Andersson B, Adamska I (2001) A chloroplast DegP2 protease
essential for Escherichia coli survival at high temperatures, is an endo-
performs the primary cleavage of the photodamaged D1 protein in plant
peptidase. J Bacteriol 172: 1791–1797
photosystem II. EMBO J 20: 713–722
Mattoo AK, Hoffman-Falk H, Marder JB, Edelman M (1984) Regulation of
Huesgen PF, Schuhmann H, Adamska I (2005) The family of Deg proteases
protein metabolism: coupling of photosynthetic electron transport to in
Plant Physiol. Vol. 152, 2010
PSII Protein Degradation by DEG7
vivo degradation of the rapidly metabolized 32-kilodalton protein of the
(2002) Proteome map of the chloroplast lumen of Arabidopsis thaliana. J
chloroplast membranes. Proc Natl Acad Sci USA 81: 1380–1384
Biol Chem 277: 8354–8365
Meurer J, Plu¨cken H, Kowallik KV, Westhoff P (1998) A nuclear-encoded
Schuster G, Timberg R, Ohad I (1988) Turnover of thylakoid photosystem
protein of prokaryotic origin is essential for the stability of photosystem
II proteins during photoinhibition of Chlamydomonas reinhardtii. Eur J
II in Arabidopsis thaliana. EMBO J 17: 5286–5297
Biochem 177: 403–410
Nelson N, Yocum CF (2006) Structure and function of photosystems I and
Silva P, Thompson E, Bailey S, Kruse O, Mullineaux CW, Robinson C,
II. Annu Rev Plant Biol 57: 521–565
Mann NH, Nixon PJ (2003) FtsH is involved in the early stages of repair
Ohad I, Kyle DJ, Arntzen CJ (1984) Membrane protein damage and repair:
of photosystem II in Synechocystis sp. PCC 6803. Plant Cell 15: 2152–2164
removal and replacement of inactivated 32-kilodalton polypeptides in
Spiess C, Beil A, Ehrmann M (1999) A temperature-dependent switch
chloroplast membranes. J Cell Biol 99: 481–485
from chaperone to protease in a widely conserved heat shock protein.
Ortega JM, Roncel M, Losada M (1999) Light-induced degradation of
Cell 97: 339–347
cytochrome b559 during photoinhibition of the photosystem II reaction
Strauch KL, Beckwith J (1988) An Escherichia coli mutation preventing
center. FEBS Lett 458: 87–92
degradation of abnormal periplasmic proteins. Proc Natl Acad Sci USA
Pallen M, Wren B (1997) The HtrA family of serine proteases. Mol
Microbiol 26: 209–221
Sun XW, Peng LW, Guo JK, Chi W, Ma JF, Lu CM, Zhang LX (2007a)
Peltier JB, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A,
Formation of DEG5 and DEG8 complexes and their involvement in the
Liberles DA, Soderberg L, Roepstorff P, von Heijne G, et al (2002)
degradation of photodamaged photosystem II reaction center D1 pro-
Central functions of the luminal and peripheral thylakoid proteome of
tein in Arabidopsis thaliana. Plant Cell 19: 1347–1361
Arabidopsis determined by experimentation and genome-wide predic-
Sun XW, Wang LY, Zhang LX (2007b) Involvement of DEG5 and DEG8
tion. Plant Cell 14: 211–236
Prasil O, Adir N, Ohad I (1992) Dynamics of photosystem II: mechanisms
proteases in the turnover of the photosystem II reaction center D1
of photoinhibition and recovery process. In J Barber, ed, The Photosys-
protein under heat stress in Arabidopsis thaliana. Chin Sci Bull 52:
tems: Structure, Function and Molecular Biology, Vol 11. Elsevier Sci-
ence Publishers, Amsterdam, pp 295–348
Wilken C, Kitzing K, Kurzbauer R, Ehrmann M, Clausen T (2004) Crystal
Sakamoto W (2006) Protein degradation machineries in plastids. Annu Rev
structure of the DegS stress sensor: how a PDZ domain recognizes
Plant Biol 57: 599–621
misfolded protein and activates a protease. Cell 117: 483–494
Sakamoto W, Zaltsman A, Adam Z, Takahashi Y (2003) Coordinated
Yamamoto Y, Akasaka T (1995) Degradation of antenna chlorophyll-
regulation and complex formation of yellow variegated1 and yellow
binding protein CP43 during photoinhibition of photosystem II. Bio-
variegated2, chloroplastic FtsH metalloproteases involved in the repair
chemistry 34: 9038–9045
cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell
Zhang LX, Paakkarinen V, van Wijk KJ, Aro EM (1999) Co-translational
assembly of the D1 protein into photosystem II. J Biol Chem 274: 16062–
Schubert M, Petersson UA, Haas BJ, Funk C, Schroder WP, Kieselbach T
Plant Physiol. Vol. 152, 2010
Source: http://sourcedb.ib.cas.cn/cn/ibthesis/201004/P020100416606476792521.pdf
Cost effective C3-fermentation chemistry The NordBioChem's way out of Fossils Cologne, April 7th 2016 NordBioChem will in following show, that • an efficient and competitive C3-fermentation-chemisry is available and next to PLA • a high variety of C3-Bio-Chemicals like propylene oxide, propylene glycol, their derivatives, acrylates as well as many other basic and new chemicals could be produced.
Medical Information Network Distribution Service Eickholz P, Dannerritz B, Kim TS. Antibiotics in periodontal therapy. Perio. 2005 Dahlen G. Microbiology and treatment of dental abscesses and periodontal endodontic lesions. Periodontol 2000. 2002 Umeda M, Tominaga Y, Yano K, Watanabe H, Ishikawa I. Microbial flora in the acute phase of periodontitis and the effect of local admini-





