Levitra enthält Vardenafil, das eine kürzere Wirkdauer als Tadalafil hat, dafür aber schnell einsetzt. Männer, die diskret bestellen möchten, suchen häufig nach levitra kaufen ohne rezept. Dabei spielt die rechtliche Lage in der Schweiz eine wichtige Rolle.
A modular positive feedback-based gene amplifier
Nistala et al. Journal of Biological Engineering 2010, 4:4http://www.jbioleng.org/content/4/1/4
A modular positive feedback-based geneamplifier
Goutam J Nistala1†, Kang Wu2†, Christopher V Rao2*, Kaustubh D Bhalerao1
Background: Positive feedback is a common mechanism used in the regulation of many gene circuits as it canamplify the response to inducers and also generate binary outputs and hysteresis. In the context of electrical circuitdesign, positive feedback is often considered in the design of amplifiers. Similar approaches, therefore, may beused for the design of amplifiers in synthetic gene circuits with applications, for example, in cell-based sensors.
Results: We developed a modular positive feedback circuit that can function as a genetic signal amplifier,heightening the sensitivity to inducer signals as well as increasing maximum expression levels without the needfor an external cofactor. The design utilizes a constitutively active, autoinducer-independent variant of the quorum-sensing regulator LuxR. We experimentally tested the ability of the positive feedback module to separately amplifythe output of a one-component tetracycline sensor and a two-component aspartate sensor. In each case, thepositive feedback module amplified the response to the respective inducers, both with regards to the dynamicrange and sensitivity.
Conclusions: The advantage of our design is that the actual feedback mechanism depends only on a single geneand does not require any other modulation. Furthermore, this circuit can amplify any transcriptional signal, not justone encoded within the circuit or tuned by an external inducer. As our design is modular, it can potentially beused as a component in the design of more complex synthetic gene circuits.
showed that positive feedback could be used to generate
Positive feedback is a common mechanism involved in
hysteresis with respect to an inducer in mammalian
the regulation of genetic circuits Any time a gene
cells. Maeda and Sano analyzed a synthetic positive
product has the capacity to enhance its own production,
feedback loop in E. coli and demonstrated that it could
either directly or indirectly, the circuit is said to involve
give rise to either a graded or hysteretic response
positive feedback. A number of behaviors can be attribu-
depending on the specific configuration. In terms of
ted to positive feedback loops. The defining one is
building circuits, Ajo-Franklin and coworkers []
clearly amplification. More complex behaviors include
demonstrated that positive feedback could be used to
bistability and hysteresis. In addition, positive feedback
engineer memory into yeast cells. Stricker and cowor-
is an integral element in many oscillatory, pattern-for-
kers on the other hand, built a simple oscillator by
mation, and intracellular polarization processes
coupling positive feedback with negative feedback. In
In a number of synthetic biology applications, positive
work most closely related to the present study, Sayut
feedback has been used to design switches, oscillators,
and coworkers [demonstrated that a positive feed-
and amplifiers. Besckei and coworkers for example,
back loop could make the transcriptional activity of the
showed in yeast that a simple positive feedback loop
quorum-sensing regulator LuxR more sensitive to auto-
could transform a graded response to an inducer into a
inducer. In these regards, their design is most closely
binary one. Likewise, Kramer and Fussenegger []
related to how positive feedback is typically employed inelectronic circuits, namely to amplify the response to asignal.
* Correspondence: † Contributed equally
In this work, we constructed a modular genetic ampli-
2Department of Chemical and Biomolecular Engineering, University of Illinois
fier in Escherichia coli based on a constitutively active,
at Urbana-Champaign, 600 S Mathews Ave, Urbana, IL, 61801, USA
2010 Nistala et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative CommonsAttribution License ), which permits unrestricted use, distribution, and reproduction inany medium, provided the original work is properly cited.
Nistala et al. Journal of Biological Engineering 2010, 4:4
autoinducer-independent variant of the quorum-sensing
Table 1 Plasmids used in this study
regulator LuxR from Vibrio fischeri Our goal was
Relevant characteristic
to develop a simple network component that could be
bla Plpp-taz ori p15A
coupled to any cell-based sensing system where the out-
cm PLtetO-1 ori ColE1
put involves the transcription of some gene. In these
kan GFP[tagless] ori p15A
regards, we sought to engineer an "off the shelf" device
that could be readily implemented in any gene circuit.
LlacO-1-luc ori ColE1
To test the ability of this device to amplify a transcrip-
kan PLtetO-1 ori ColE1
tional output, we coupled our device to a one-compo-
kan Plac/ara-1 ori pSC101
nent tetracycline sensor and a two-component aspartate
cm Plac/ara-1-luxR-luxI ori ColE1
sensor. In both cases, we found that our amplifier was
kan PLtetO-1 ori p15A
able to increase the sensitivity to the input signal and
kan PLtetO-1-luxR * ori p15A
intensify the output signal.
kan PLtetO-1-luxRΔ2-156 ori p15A
kan PLtetO-1-luxRΔ2-162 ori p15A
cm Plux ori ColE1
Media, growth conditions, and bacterial strains
All cultures experiments were performed in either
OmpC-luxRΔ2-162 ori p15A
Luria-Bertani (LB) broth (tryptone: 10 g/L, yeast extract:
lux -GFP [tagless]-luxRΔ2-162 ori
5 g/L, and NaCl: 10 g/L) or M9 minimal media supple-
cm Plux-GFP[tagless] ori ColE1
ment with 0.4% glucose, 1 μg/mL thiamine, and 1 μg/
mL biotin. All experiments were performed at 37°C
LtetO-1 ori ColE1
unless noted otherwise. Antibiotics were used at the fol-
bla PLtetO-1-taz ori ColE1
lowing concentrations: ampicillin at 100 μg/mL, chlor-
bla PLtetO-1-taz ori pSC101
amphenicol at 20 μg/mL, and kanamycin at 40 μg/mL.
Plasmids are from this study unless noted otherwise.
Primers were purchased from IDT Inc. (Coralville, IA).
Restriction enzymes were purchased from New EnglandBiolabs Inc. (Ipswitch, MA) and Fermentas Inc. (Glen
origin from pZA34-luc using the restriction sites XbaI
Burnie, MD) and used according to the manufacturer's
and SacI and by swapping the chloramphenicol resis-
tance gene with the kanamycin resistance gene from
All cloning steps were performed in E. coli strain
pZE21 using the restriction sites XhoI and SacI. The
DH5a. Subsequent experiments involving anhydrotetra-
plasmid pPROTetE-Amp was made by replacing
cycline induction were conducted in E. coli strain
the chloramphenicol resistance gene in pPROTet.E with
GN100 (F- ilvG rfb-50 rph-1 ΔenvZ::FRT attBl::[PN25-
the ampicillin resistance gene from pZE12 using the
tetR lacIq spcR]) and those involving aspartate induction
restriction sites XhoI and SacI.
were performed in GN101 (F- ilvG rfb-50 rph-1 ΔenvZ::
The luxI-GFP transcriptional fusion was made first by
FRT). Strain GN100 was constructed first by P1vir
PCR amplification of the luxI promoter using the plasmid
transduction of the ΔenvZ::kan insert from JW3367-3
pluxGFPuv [as the template with the primers
(The E. coli Genetic Stock Center, CGSC# 10509) into
KW134F (CAG ATA TCG ACG TCA GTC C) and
MG1655. The antibiotic cassette from the FRT-Kan-
KW134R2 (ATA GAA TTC TGC GTT TAT TCG ACT
FRT insert was then removed by transformation of
ATA AC). The resulting fragment was then cloned into
pCP20 into the strain and selection on ampicillin at
the plasmid pPROTet.E using the restriction sites EcoRI
30°C . Loss of the helper plasmid pCP20 was
and AatII, yielding the plasmid pGN23. The green fluor-
obtained by growth at 42°C under non-selective condi-
escent protein (GFP) was PCR amplified from pPROBE-
tions on LB agar. Lastly, the chromosomally integrated
gfp[tagless] ] using primers GN10F (GGG GAA TTC
TetR/LacI expression cassette from DH5aZ1 was
ATA CGT ATT TAA ATC AGG AGT GGA AAT GAG
moved into this strain by P1vir transduction, yielding
TAA AGG AGA AGA ACT T) and GN10R (GGG GGA
GN100. Similarly, strain GN101 was constructed in an
TCC TTA TTA TTT GTA TAG TTC ATC CA). The
identical manner except that it does not harbor the
resulting fragment was then cloned into the EcoRI and
TetR/LacI expression cassette from DH5aZ1.
BamHI restriction sites of the pGN23, yielding the plas-mid pGN69.
Plasmids Construction
The LuxR* (LuxR[A221V]) expression plasmids were
Table provides a list of the plasmids used in this
constructed using two rounds of PCR. In the first
study. The plasmid pPROTetE-Kan-p15A was made by
round, the luxR gene was amplified with primers
swapping the ColE1 origin of pPROTet.E with the p15A
KW78F1 (AAC TTT ATA AGG AGG AAA AAC ATA
Nistala et al. Journal of Biological Engineering 2010, 4:4
TGA AAA ACA TAA ATG CCG AC) and KW078R
TGT TAT TAA CCC). The PCR product was then
(ACT GTC GAC TTA ATT TTT AAA GTA TGG GC)
digested with XhoI and EcoRI and sub-cloned into the
using pLuxRI [as the template. The resulting pro-
respective sites of pPROTetE-Kan-p15A, thus replacing
duct was then used as a template for a second round of
the native PLtetO-1 promoter with the PompC promoter.
PCR this time using primers KW078F2 (TAT GAA
The primers GN06F2 (GGG GTC GAC ATG CCT TCT
TTC AAC TAA AGA TTA ACT TTA TAA GGA GGA
CTA GTT GAT AA) and KW171R were used to
AAA ACA) and KW078R. It was then digested with
amplify luxRΔ2-162 using pGN68 as the template. The
EcoRI and SalI and sub-cloned into the EcoRI and SalI
resulting PCR product was digested with SalI and NotI
cut-sites of pPROTetE-Kan-p15A. Enzymatic inverse
and then sub-cloned into the respective sites of pPRO-
PCR was used to introduce the Ala221Val (GCG- >
TetE-Kan-p15A, yielding pGN62.
GTG) point mutation in the luxR gene with primers
The aspartate sensor module was constructed first
KW079F (ATA GGT CTC TGT GCA AAT GAA ACT
amplifying the taz gene from pTJ003 using the primers
CAA TAC AAC) and KW079R (ATA GGT CTC TGC
GN13F (GGG GAA TTC TTA AAG AGG AGA AAG
ACA TTG GTT AAA TGG AAA GTG A). The result-
GTA CCC ATG ATT AAC CGT ATC C) and GN12R
ing PCR product was then digested with BsaI and
(GGG GTC GAC TTA CCC TTC TTT TGT CGT
ligated to obtain pGN3.
GCC CT). The PCR product was then digested with
The luxRΔ2-156 expression plasmid was also con-
EcoRI and SacI and cloned into the unique respective
structed using two rounds of PCR. The luxR gene was
restriction sites, yielding pGN76. The ColE1 origin in
first amplified with primers KW112F (AAC TTT ATA
pGN76 was then replaced with the pSC101 origin from
AGG AGG AAA AAC ATA TGA ACA TAC CAT
the pZS24 plasmid using the restriction sites AvrII and
TAA TTG TTC C) and KW078R using pLuxRI as the
SacI, yielding pGN77.
template. The resulting PCR product was then amplifiedusing primers KW078F2 and KW078R. It was then
Fluorescence Assays
cloned into the EcoRI and SalI cut-sites of pPROTetE-
To measure fluorescent protein expression, cultures
Kan-p15A, yielding pGN11. Likewise, the luxRΔ2-162
were first grown overnight and then subcultured to an
expression plasmid was made by amplifying the luxR
OD600 of 0.05 in fresh media. The cultures were first
gene with primers KW113F (CTT TAT AAG GAG
allowed to grow to an OD600 of 0.20, at which point the
GAA AAA CAT ATG CCT TCT CTA GTT GAT AAT
inducer was added. The cultures were then grown over-
TAT C) and KW078R using pLuxRI as the template.
night prior to taking the measurements. 100 μL of the
The resulting product was amplified again as before
culture was then transferred into a 96 well microplate,
using primers KW078F2 and KW078R. The PCR
and the relative fluorescence and optical density at
product was then digested with EcoRI and SalI and sub-
600 nm (OD600) were measured using a Tecan Safire2
cloned into the EcoRI and SalI cut-sites of pPROTetE-
microplate reader. The fluorescence readings, given as
Kan-p15A, yielding pGN12.
relative fluorescence units (RFU), were normalized with
The positive-feedback module was constructed using
the OD600 absorbance to account for cell density. All
two rounds of PCR. In the first round, the primers
experiments were performed in triplicate with 95% con-
GN09F2 (AAC TAA AGA TTA ACT TTA TAA GGA
fidence intervals reported.
GGA AAA ACA TAT GCC TTC TCT AGT TGA TAAT) and KW171R (AAT AGC GGC CGC TTA TTA
Results and Discussion
ATT TTT AAA GTA TGG GC) were used to amplify
Design of positive-feedback amplifier
the luxRΔ2-162 domain using pLuxRI [as the
In order to construct a positive feedback circuit, we
template. The resulting PCR product was then used as
required a transcriptional activator that did not interfere
template for a second round of PCR this time using pri-
with native gene regulation in E. coli. In addition, we
mers GN09F (GGG GGA TCC AAC TAA AGA TTA
required that the activator be constitutively active and
ACT TTA TAA GGA GGA AAA ACA T) and
not dependent on the addition of an exogenous inducer.
KW171R (AAT AGC GGC CGC TTA TTA ATT TTT
Given these constraints, we chose the LuxR protein
AAA GTA TGG GC). The resulting fragment was then
from Vibrio fischeri This protein, normally involved
digested with BamHI and NotI and sub-cloned into
in quorum sensing and bioluminescence, activates the
pGN69, yielding pGN68.
transcription of the luxIADCBE operon in response to
The aspartate positive feedback module was con-
acyl homoserine lactone (AHL). AHL binding stabilizes
structed first by amplifying the PompC promoter (geno-
the LuxR dimer and, as a result, increases its ability to
mic region 2310762-2310962) using primers GN03F
activate transcription
(GGG CTC GAG GTT CCC TTG CAT TTA CAT
While wild-type LuxR does not appear to interfere
TTT) and GN05R (GGG GAA TTC TAA CTT TCA
with native E. coli regulation, it still requires an
Nistala et al. Journal of Biological Engineering 2010, 4:4
exogenous inducer. However, a number of approaches
relative to the wild-type control. Based on these results,
exist for making constitutively active derivatives of LuxR
we chose to use the LuxRΔ2-162 variant to design the
and thus satisfying our design constraints. For example,
an Ala221Val point mutation was previously found to
To construct the amplifier, we cloned GFP and
constitutively activate LuxR . The alanine at position
LuxRΔ2-162 in a bicistronic configuration behind the
221 enables the N-terminal signaling domain to inhibit
PluxI promoter on high-copy number plasmid (ColE1
the activity of the C-terminal, DNA-binding domain.
origin of replication). In this arrangement, LuxRΔ2-162
Presumably, mutating this residue to a valine prevents
functions in a positive feedback loop as it can bind to
the N-terminal domain from interfering with DNA
the PluxI promoter and activate its own transcription
binding. Consistent with this model, deleting the N-
(Figure ). The reason we cloned LuxRΔ2-162 down-
terminal domain of LuxR was also found to yield a con-
stream of the GFP reporter is to control for polar effects
stitutively active variant
when we compared results involving positive feedback
Based on these previous studies, we engineered three
to those lacking it. To induce this circuit, we again used
constitutively active variants of LuxR to test their suit-
LuxRΔ2-162, this time as the input signal. In such a
ability in designing an amplifier. The first, denoted by
design, the output of the sensor is LuxRΔ2-162, which in
LuxR*, harbors the Ala221Val point mutation. The
turn feeds back into the amplifier. In these regards,
other two, denoted by LuxRΔ2-156 and LuxRΔ2-162
LuxRΔ2-162 is used both as the input and positive feed-
respectively, involved different N-terminal deletions,
back signal. For the output, we used GFP as it provides
where the subscript denotes the deleted fragment. To
a facile measure of transcriptional activity. This choice
test the relative effectiveness of these three different
is in no way limiting, and any gene can in practice be
constitutive LuxR variants, we determined how strongly
used as the output.
they could activate expression from the PluxI promoter,using the green fluorescent protein (GFP) as our tran-
Validation of amplifier using a tetracycline sensor
scriptional readout. The results from these experiments
We first tested the amplifier by coupling it to a one-
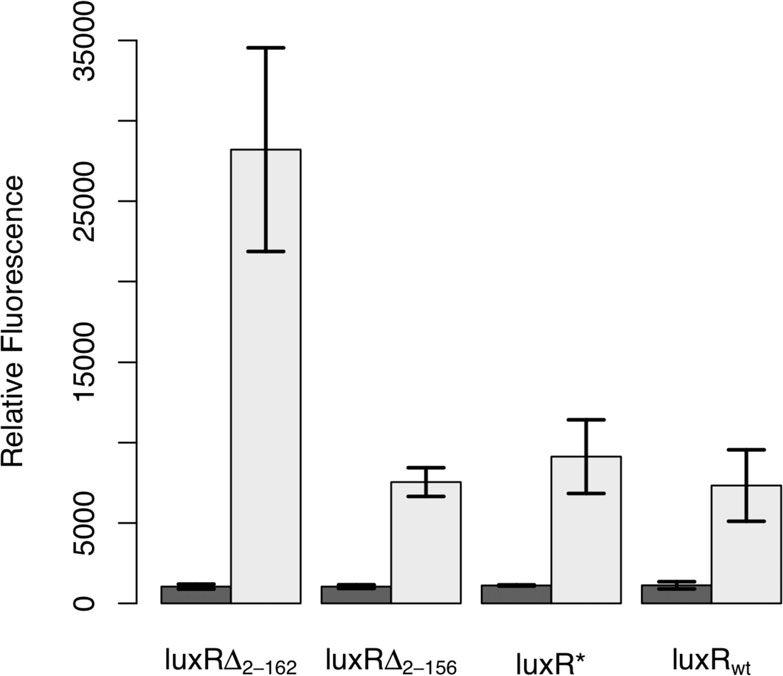
are shown in Figure All of the LuxR variants, includ-
component tetracycline sensor (Figure ). In this design,
ing the wild-type control, were able to induce expression
we cloned LuxRΔ2-162 behind the TetR-regulated PLtetO-1
from the PluxI promoter. Of the three, only LuxRΔ2-162
promoter on a compatible, medium copy-number plas-
was capable in our hands of enhancing transcription
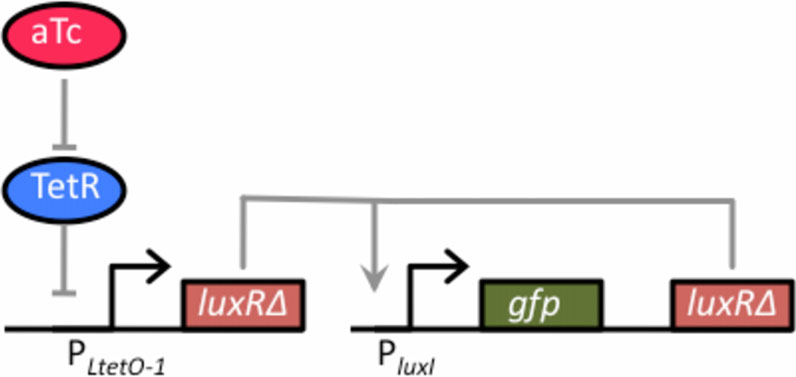
mid (p15A origin of replication) In the absence ofthe tetracycline analogue, anhydrotetracycline (aTc),dimeric TetR binds to the O2 operator sites within thePLtetO-1 promoter and represses transcription. However,when TetR is bound with aTc, it no longer binds andrepresses the PLtetO-1 promoter, enabling dose-depen-dent control of gene expression. Thus, the aTc-induciblepromoter functions as a one-component tetracyclinesensor with LuxRΔ2-162 as the output.
To couple this sensor with the amplifier, we trans-
formed cells (GN100) constitutively expressing a
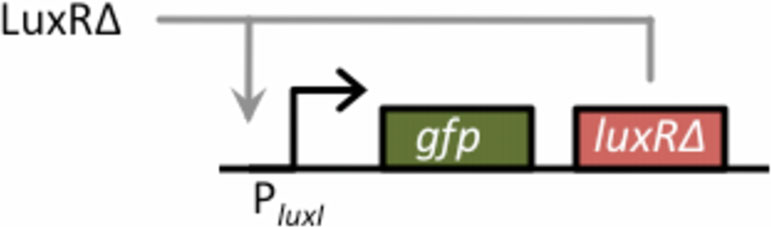
Figure 2 Schematic of positive-feedback amplifier. The basic
Figure 1 Comparison of constitutive LuxR variants. In these
design for the amplifier consists of GFP and LuxRΔ2-162 arranged in
experiments, LuxR was expressed from a tetracycline-inducible
a bicistronic configuration under the control of the PluxI promoter.
promoter, PLtetO-1, in strain GN101, which harbors a chromosomal
LuxRΔ2-162 functions in a positive feedback loop as it can bind to
copy of tetR. Activity was determined by the ability of these
the PluxI promoter and activate its own transcription. In our design,
different variants to induce expression from the PluxI promoter,
LuxRΔ2-162 is also used as the input signal for the amplifier. LuxRΔ2-
using GFP as the readout, in the absence of any autoinducer. Dark
162, therefore, functions both as the input and positive feedback
bars denote the uninduced case and light bars the induced case
signal. GFP, the output signal, provides a measure of transcriptional
(200 ng/mL aTc). Error bars denote 95% confidence intervals.
Nistala et al. Journal of Biological Engineering 2010, 4:4
Figure 3 Schematic of tetracycline sensor coupled to thepositive-feedback amplifier. The one-component tetracyclinesensor consists of a plasmid where LuxRΔ2-162 has been clonedbehind the TetR-regulated PLtetO-1 promoter. In the absence of theinducer anhydrotetracycline (aTc), dimeric TetR binds to the O2operator sites within the PLtetO-1 promoter and repressestranscription. However, when bound with aTc, TetR is no longerable to bind to the O2 operator sites within the promoter, thusenabling dose-dependent control of LuxRΔ2-162. This sensor wascoupled with the positive feedback amplifier, encoded on a
Figure 4 Comparison of tetracycline sensor with positive
separate plasmid, by transforming cells (GN100) constitutively
feedback (solid circles) and without (solid square). Schematic of
expressing a chromosomal copy of the tetR gene with the two
positive feedback design is shown in Figure 3. The design lacking
plasmids respectively harboring the sensor and amplifier.
positive feedback is otherwise identical to one with positivefeedback except that only GFP is expressed from the PluxI promoter.
In these experiments, cells were grown overnight at the indicated
chromosomal copy of the tetR gene with the two plas-
concentrations of aTc prior to measurements. The fluorescencevalues were normalized with the OD
mids respectively harboring the sensor and amplifier
600 absorbance to account for
cell density. Error bars denote 95% confidence intervals for the
(see Materials and Methods for details). A schematic
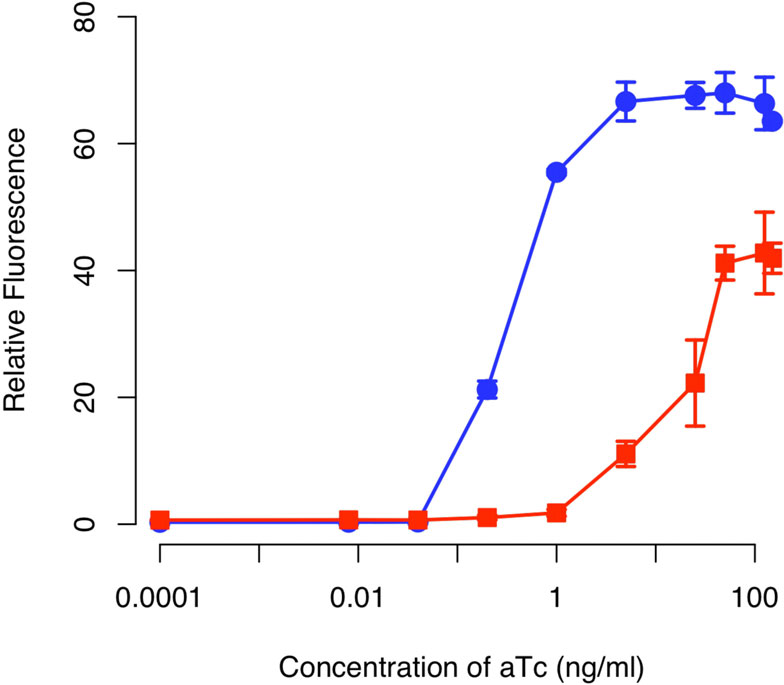
of the integrated design is given in Figure When wetested this design, we found that the amplifierincreased both the sensitivity and dynamic range of
multiple studies have shown that positive feedback can
the integrated circuit relative to an otherwise identical
lead to bistability and hysteresis Therefore, we
circuit lacking positive feedback (Figure ). In particu-
speculated that cells harboring the amplifier might be
lar, we found that positive feedback increased the sen-
able to "remember" previous exposures to aTc. How-
sitivity to aTc by roughly two orders of magnitude. In
ever, when we transferred cells from media containing
other words, we observed equivalent levels of expres-
aTc to media lacking it, we no longer observed any
sion in the circuit involving positive feedback at aTc
GFP expression relative to the background after we
concentrations roughly one hundred times less than
grew the cells up (data not shown). These results indi-
those observed with the circuit lacking positive feed-
cate the positive feedback loop involving LuxRΔ2-162 is
back. Moreover, we found that positive feedback
able to amplify the response to an inducer but is
increased the dynamic range by roughly 50%. By range,
incapable of sustaining the response in the absence of
we mean the ratio of expression under saturating indu-
cing (100 ng/ml aTc) and non-inducing (0 ng/ml aTc)
Based on what we know about the properties of
LuxR, specifically the role of AHL in stabilizing LuxR,
In addition to these endpoint measurements, we also
the reason the circuit does not sustain activation is
performed kinetic experiments where we measured the
likely due to the protein dimer being degraded too
response over a twelve-hour interval to varying concen-
quickly. In other words, we suspect that LuxRΔ2-162
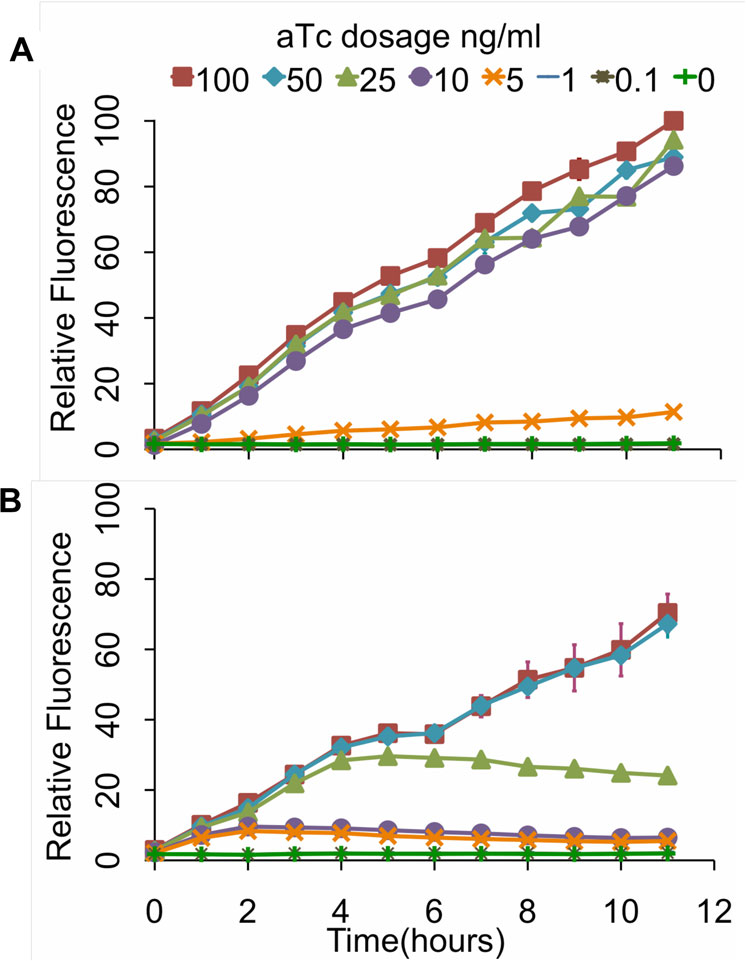
trations of aTc (Figure Consistent with our end-
dimer is being degraded at a rate greater than it is
point measurements, we found that the design involving
being produced by positive feedback alone (though we
positive feedback was more sensitive to aTc and had a
did not directly make this measurement). More specifi-
wider dynamic range of expression levels. Collectively,
cally, positive feedback alone is unable to sustain the
these results demonstrate that our genetic amplifier is
expression of LuxRΔ2-162 in the absence of some exo-
capable of both increasing the sensitivity and dynamic
genous source, in our case the one-component sensor.
range of this one-component tetracycline sensor.
That said, the positive feedback is still strong enough
We last tested whether the amplifier would endow
to amplify the response when an external input signal
the cell with memory. While not a design goal,
Nistala et al. Journal of Biological Engineering 2010, 4:4
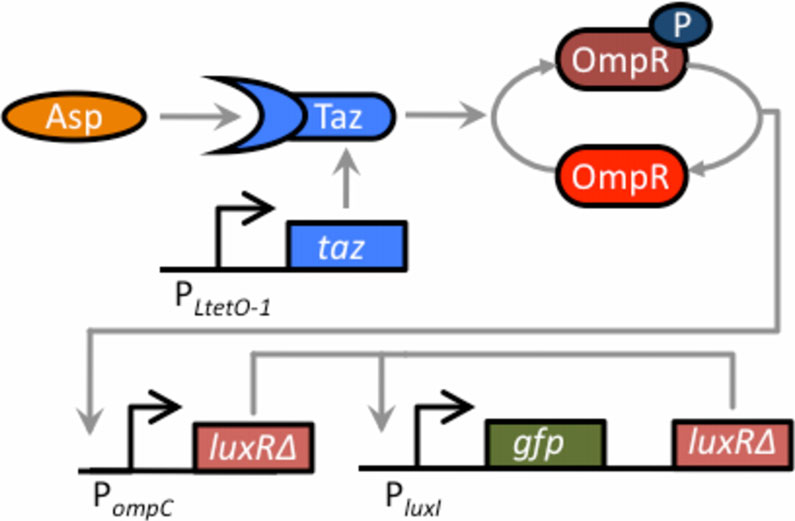
Figure 6 Schematic of aspartate sensor coupled to thepositive-feedback amplifier. The two-component sensor consistsof the Taz sensor kinase and the OmpR response regulator. Tazcontrols the level of phosphorylated OmpR (OmpR-P), which in turnactivates the expression from the PompC promoter. When the Tazsensor kinase is bound with aspartate, it increases the levels ofOmpR-P, leading to increased expression from the PompC promoter.
In our design, the Taz sensor kinase has been cloned behind theconstitutive PLtetO-1 promoter on one plasmid (the cells used inthese experiments do not possess TetR). On a second plasmid,LuxRΔ2-162 has been cloned behind the PompC promoter, resulting inthe expression of LuxRΔ2-162 being aspartate dependent. The thirdplasmid harbors the positive feedback amplifier. The sensor was
Figure 5 Kinetic analysis of tetracycline sensor with positive
coupled to the amplifier by transforming the three plasmids into a
feedback (A) and without (B). In these experiments, cells were
ΔenvZ null mutant (GN101).
grown for 12 hours at varying levels of aTc induction withmeasurements taken every hour. The fluorescence values werenormalized with the OD600 absorbance to account for cell density.
The scale for both sets of experiments is the same. Error bars
To construct the integrated circuit in E. coli, we trans-
denote 95% confidence intervals for the measurement average.
formed a ΔenvZ null mutant (GN101) with these threeplasmids.
Similar to what we observed with the one-component
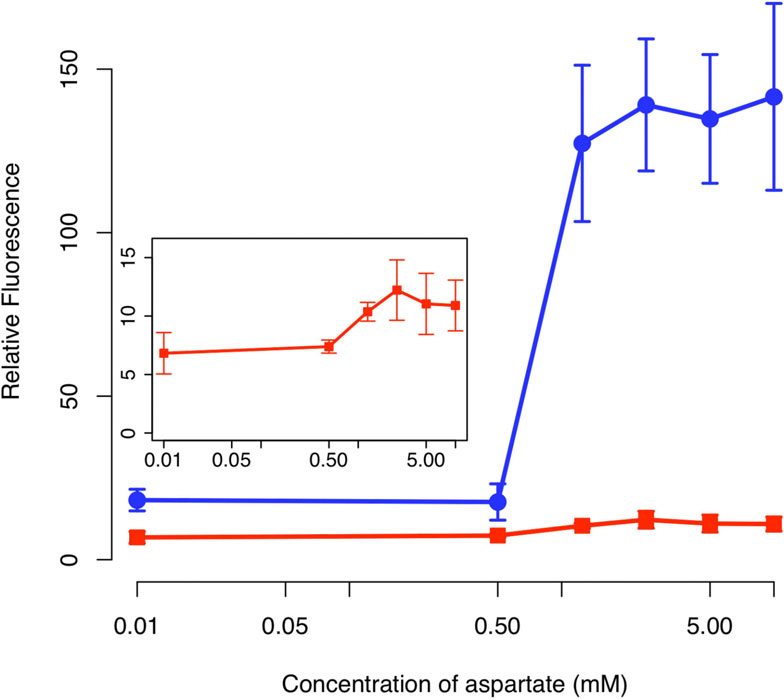
Validation of amplifier using an aspartate sensor
tetracycline receptor, we found that the amplifier
We next tested the amplifier by coupling it to a two-
increased both the range and sensitivity when coupled
component aspartate sensor (Figure To do this, we
to the two-component aspartate sensor as compared to
used the hybrid Tar-EnvZ (Taz) sensor kinase [. This
an otherwise identical circuit lacking positive feedback
chimeric, transmembrane sensor kinase controls the
(Figure Unlike the case with the one-component sen-
levels of phosphorylated OmpR, which in turn activates
sor, we observed only a minor increase in sensitivity.
the expression from the PompC promoter. When the Taz
However, we observed a significant amplification of the
sensor kinase is bound with aspartate, it increases the
response. In particular, the amplifier increased the
levels of OmpR-P, leading to increased expression from
dynamic range by roughly an order of magnitude
the PompC promoter. In addition to amino acids, EnvZ
whereas the sensitivity increased by approximately a fac-
chimeras have been constructed to sense other inputs
tor of two. While these results demonstrate that the
such as sugars and light
amplifier is modular as it can readily be applied to dif-
In order to couple the two-component aspartate sen-
ferent sensor systems, they also demonstrate that the
sor with our genetic amplifier, we cloned LuxRΔ2-162
performance of the amplifier is context dependent. In
behind PompC promoter on a compatible, medium copy-
particular, we observed mostly an increase in the range
number plasmid (p15A origin of replication). To intro-
when the amplifier was coupled to the two-component
duce the Taz sensor kinase into E. coli, we cloned this
aspartate sensor kinase and, conversely, mostly an
gene behind the constitutive PLtetO-1 promoter on a
increase in sensitivity when it was coupled to the one-
compatible, low copy-number plasmid (pSC101 origin of
component tetracycline sensor.
replication). Note, these experiments were performed in
We note that we observed only weak activation,
cells lacking a chromosomal copy of the tetR gene, so
roughly two-fold, in response to aspartate in the absence
the PLtetO-1 promoter in this background is constitutive.
of positive feedback. This level of activation is less than
Nistala et al. Journal of Biological Engineering 2010, 4:4
In addition to sensing applications, the amplifier can
also be used to create devices of greater complexity infunction. One intriguing application concerns impe-dance matching. Impedance mismatch occurs when theoutput range of one sub-circuit does not match theinput range of another sub-circuit to which it is con-nected. To effectively link these two sub-circuits, therespective output and input ranges should match oneanother. As positive feedback can significantly alter theresponse of a sub-circuit, it can be used as an ‘impe-dance matching' device by coupling two different sub-circuit circuits together that have disparate requirementsfor signal levels to operate correctly.
A primary goal of synthetic biology is to design modu-
lar components with defined behavior that can be reusedin diverse applications The ideal componentshould have predicable behavior regardless of the contextin which it is applied. This is a significant challenge. Even
Figure 7 Comparison of sensor output in the presence (solid
in our experiments, while we rightly hypothesized that
circles) and absence (solid squares) of the positive feedbackamplifier. Schematic of positive feedback design is shown in
we would see amplification due to the positive feedback,
Figure 6. The design lacking positive feedback is otherwise identical
we see a different response when we coupled the ampli-
to one with positive feedback except that only GFP is expressed
fier to the two different sensors. For instance, the tetracy-
from the PluxI promoter. In these experiments, cells were grown
cline sensor showed a major increase in sensitivity but
overnight at the indicated concentrations of aspartate prior to
only moderate increase in the dynamic response. The
measurements. The fluorescence values were normalized with theOD
aspartate sensor, however, showed a major increase in
600 absorbance to account for cell density. Inset figure shows the
magnification of the response for the design lacking positive
the dynamic response but only a moderate increase in
feedback. Error bars denote 95% confidence intervals for measured
sensitivity. Moreover, the amplifier increased background
expression in the case of the aspartate sensor but not inthe case of the tetracycline sensor. The origins of these
what has been previously observed in other studies
differences are unknown, but may arise due to variations,
using Taz, where the degree of activation is greater than
for example, in plasmid copy number, promoter
ten fold However, unlike our design, these stu-
strengths, and the metabolic burden imposed by each cir-
dies measured the expression from the PompC promoter.
cuit. While further engineering can be used to control for
In the present work, we measured the expression from a
these individual factors, their effects are often non-trivial
downstream promoter, PluxI. Thus, there is an additional
to isolate and quantify.
stage between the sensor and reporter in our design.
Likely, expression of LuxRΔ2-162 from the PompC promo-ter is not sufficiently strong to activate the P
We thank Lingchong You and Christopher Voigt for plasmids. This work was
ter without further amplification. However, when we
supported by National Science Foundation grants 0644744 (to CR) and
add amplification by including positive feedback, we
0943386 (to KB), as well as start up funds provided by the University of
then obtain robust expression.
Illinois (to KB).
1Department of Agricultural and Biological Engineering, University of Illinois
In this work, we developed a simple modular genetic
at Urbana-Champaign, 1304 W Pennsylvania Ave, Urbana, IL, 61801, USA.
2Department of Chemical and Biomolecular Engineering, University of Illinois
amplifier based on a constitutively active variant of LuxR.
at Urbana-Champaign, 600 S Mathews Ave, Urbana, IL, 61801, USA.
We tested this amplifier by coupling it to a one-compo-nent tetracycline sensor and a two-component aspartate
Authors' contributionsCR and KB conceived the experiments. GN and KW performed experiments.
sensor. In both instances, the amplifier was able to
CR and KB wrote the manuscript. All authors read and approved the final
increase the dynamic range and sensitivity of the inte-
grated circuit. Based on these results, this amplifier most
Competing interests
likely can be coupled to any cell-based sensor where the
The authors declare that they have no competing interests.
output involves the transcription of a gene. In theseregards, we have successfully constructed a reusable
Received: 7 September 2009 Accepted: 26 February 2010Published: 26 February 2010
Nistala et al. Journal of Biological Engineering 2010, 4:4
Michalodimitrakis KM, Sourjik V, Serrano
Mitrophanov AY, Groisman
Mol Microbiol 2005, 58:257-266.
Bioessays 2008, 30:542-555.
Yoshida T, Phadtare S, Inouye M
Onsum MD, Rao CV:
Methods Enzymol 2007, 423:166-183.
Curr Opin Cell Biol 2009,
Trends Biotechnol 2009, 27:368-374.
Novak B, Tyson Nat Rev
Canton B, Labno A, Endy
Mol Cell Biol 2008, 9:981-991.
Nat Biotechnol 2008, 26:787-793.
Becskei A, Seraphin B, Serrano L:
Endy D: Nature 2005, 438:449-453.
Lucks JB, Qi L, Whitaker WR, Arkin AP:
Embo J 2001, 20:2528-2535.
Curr Opin Microbiol 2008,
Kramer BP, Fussenegger
Proc Natl Acad Sci USA 2005, 102:9517-9522.
J Mol Biol 2006, 359:1107-1124.
Ajo-Franklin CM, Drubin DA, Eskin JA, Gee EP, Landgraf D, Phillips I,
Silver PA: Genes Dev 2007,21:2271-2276.
Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty
Cite this article as: Nistala et al.: A modular positive feedback-based
Nature 2008, 456:516-519.
gene amplifier. Journal of Biological Engineering 2010 4:4.
Sayut DJ, Niu Y, Sun L: ACS Chem Biol 2006, 1:692-696.
Sayut DJ, Kambam PK, Sun LBiochem Biophys Res Commun 2007, 363:667-673.
Fuqua C, Greenberg EP: Nat Rev Mol Cell Biol 2002, 3:685-695.
Datsenko KA, Wanner BL: Proc Natl Acad Sci USA 2000,97:6640-6645.
Lutz R, Bujard Nucleic Acids Res 1997, 25:1203-1210.
You L, Cox RS, Weiss R, Arnold FNature 2004, 428:868-871.
Miller WG, Leveau JH, Lindow SE: Mol Plant Microbe Interact 2000,13:1243-1250.
Stevens AM, Dolan KM, Greenberg EP: Proc Natl Acad Sci USA 1994, 91:12619-12623.
Smith C, Song H, You L: J Mol Biol 2008, 382:1290-1297.
Urbanowski ML, Lostroh CP, Greenberg EP: J Bacteriol 2004,186:631-637.
Zhu J, Winans SC: Proc Natl Acad Sci USA 2001, 98:1507-1512.
Poellinger KA, Lee JP, Parales JV Jr, Greenberg EFEMS Microbiol Lett 1995, 129:97-101.
Choi SH, Greenberg EProc Natl Acad Sci USA 1991, 88:11115-11119.
Choi SH, Greenberg EJ Bacteriol 1992, 174:4064-4069.
Yao G, Lee TJ, Mori S, Nevins JR, You L: Nature cell biology 2008, 10:476-482.
Submit your next manuscript to BioMed Central
Utsumi R, Brissette RE, Rampersaud A, Forst SA, Oosawa K, Inouye M:
and take full advantage of:
Science 1989, 245:1246-1249.
• Convenient online submission
Baumgartner JW, Kim C, Brissette RE, Inouye M, Park C, Hazelbauer GL:
• Thorough peer review
• No space constraints or color figure charges
J Bacteriol 1994,176:1157-1163.
• Immediate publication on acceptance
Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M,
• Inclusion in PubMed, CAS, Scopus and Google Scholar
Davidson EA, Scouras A, Ellington AD, Marcotte EM, Voigt
• Research which is freely available for redistribution
Submit your manuscript at
Source: http://users.encs.concordia.ca/~kharma/COEN691A/Lectures8-end/Lecture9/3.Amplifer.pdf
Cerebral Cortex Advance Access published August 13, 2014 The Fault Lies on the Other Side: Altered Brain Functional Connectivity in PsychiatricDisorders is Mainly Caused by Counterpart Regions in the Opposite Hemisphere Jie Zhang1,2,†, Keith M. Kendrick3,†, Guangming Lu4,2 and Jianfeng Feng1,2,5 1Centre for Computational Systems Biology, Fudan University, Shanghai 200433, PR China, 2Fudan University – Jinling HospitalComputational Translational Medicine Center, Jinling Hospital, Nanjing University School of Medicine, Nanjing 210002, PR China,3Key Laboratory for Neuro Information, University of Electronic Science and Technology of China, School of Life Science andTechnology, Chengdu 610054, PR China, 4Department of Medical Imaging, Jinling Hospital, Nanjing University School ofMedicine, Nanjing 210002, PR China and 5Department of Computer Science, University of Warwick, Coventry, UK
HIGHLIGHTS OF PRESCRIBING INFORMATION These highlights do not include all the information needed to use TIROSINT safely and effectively. • Patients unable to swallow a capsule, including young children (generally under 6 years of age) (4) See full prescribing information for TIROSINT. • Acute myocardial infarction (4) TIROSINT (levothyroxine sodium) capsules, for oral use







