Levitra enthält Vardenafil, das eine kürzere Wirkdauer als Tadalafil hat, dafür aber schnell einsetzt. Männer, die diskret bestellen möchten, suchen häufig nach levitra kaufen ohne rezept. Dabei spielt die rechtliche Lage in der Schweiz eine wichtige Rolle.
Ita-suomenbiopankki.fi
Copyright 2002 The American Society for Pharmacology and Experimental Therapeutics
Mol Pharmacol 62:366–378, 2002
Printed in U.S.A.
Modulation of Mouse and Human Phenobarbital-ResponsiveEnhancer Module by Nuclear Receptors
JANNE M ¨AKINEN, CHRISTIAN FRANK, JOHANNA JYRKK ¨ARINNE, JUKKA GYNTHER, CARSTEN CARLBERG, andPAAVO HONKAKOSKI
Departments of Pharmaceutics (J.K., J.J., P.H.), Biochemistry (C.F., C.C.), and Pharmaceutical Chemistry (J.G.), University of Kuopio,Kuopio, Finland
Received October 24, 2001; accepted May 1, 2002
This article is available online at http://molpharm.aspetjournals.org
ABSTRACT
The constitutive androstane receptor (CAR) regulates mouse
suppress PBREM function via a coactivator-dependent pro-
and human
CYP2B genes through binding to the direct re-
cess that may have relevance in vivo. In competition experi-
peat-4 (DR4) motifs present in the phenobarbital-responsive
ments, mouse PBREM is clearly more selective for CAR than
enhancer module (PBREM). The preference of PBREM ele-
human PBREM. Pregnane X, vitamin D, and thyroid hormone
ments for nuclear receptors and the extent of cross-talk be-
receptors can potentially compete with human CAR on human
tween CAR and other nuclear receptors are currently unknown.
PBREM. In contrast to the selective nature of PBREM,
CYP3A
Our transient transfection and DNA binding experiments indi-
enhancers are highly and comparably responsive to CAR, preg-
cate that binding to DR4 motifs does not correlate with the
nane X receptor, and vitamin D receptor. In addition, the ligand
activation response and that mouse and human PBREM are
specificities of human and mouse CAR were defined by mam-
efficiently ‘insulated' from the effects of other nuclear receptors
malian cotransfection and yeast two-hybrid techniques. Our
despite their substantial affinity for DR4 motifs. Certain nuclear
results provide new mechanistic explanations to several previ-
receptors that do not bind to DR4 motifs, such as peroxisome
ously unresolved aspects of
CYP2B and
CYP3A gene regula-
proliferator-activated receptor-␣ and farnesoid X receptor, can
Phenobarbital (PB) and many structurally unrelated xeno-
Cyp2b10, human
CYP2B6, and rat
CYP2B1 and
CYP2B2
biotics induce same drug- and carcinogen-metabolizing cyto-
genes (reviewed by Honkakoski and Negishi, 2000). The
chrome P450 and other genes as a protective response di-
PBREM contains two CAR/RXR heterodimer binding sites,
rected toward elimination of these xenobiotics from the body.
NR1 and NR2, that conform to the direct repeat-4 (DR4)
Among tens of PB-inducible genes,
CYP2B genes are the
motif. Successive mutations of DR4 motifs result in gradual
most efficiently activated (reviewed by Waxman, 1999;
loss and, finally, abolition of
trans-activation by CAR in
Honkakoski and Negishi, 2000). Recent studies have estab-
HEK293 cells and induction in primary hepatocytes. It is
lished that the constitutive androstane receptor (CAR,
known that NR1 sites alone are sufficient for CAR respon-
NR1I3) is crucial for induction of
CYP2B genes by PB and
siveness (Sueyoshi et al., 1999), whereas the nuclear factor 1
(NFI) binding site between NR1 and NR2 may contribute to
cause CYP2B mRNA inducibility is lost in
CAR null mice
the full inducibility (Honkakoski et al., 1998; Kim et al.,
(Wei et al., 2000). After forming a heterodimer with retinoid
X receptor (RXR, NR2B), the xenobiotic-activated CAR binds
Several nuclear receptors (NRs), such as vitamin D recep-
to the phenobarbital-responsive enhancer module (PBREM)
tor (VDR, NR1I1), thyroid hormone receptors ␣/ (TR,
or unit located in the upstream regions of the mouse
NR1A1/2), retinoic acid receptors ␣//␥ (RAR, NR1B1/2/3),liver X receptors ␣/ (LXR, NR1H3/2), pregnane X receptor
This study was supported by Academy of Finland grants 44040 and 51610
(to P.H.) and 50331 (to C.C.).
(PXR, NR1I2) and farnesoid X receptor (FXR, NR1H4) dis-
ABBREVIATIONS: PB, phenobarbital; CAR, constitutive androstane receptor; TCPOBOP, 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene; RXR,
retinoid X receptor; PBREM, phenobarbital-responsive enhancer module; DR
n, direct repeat with
n base-pair spacing; ER
n, everted repeat with
n base-pair spacing; HEK, human embryonic kidney; NR, nuclear receptor; NFI, nuclear factor 1; VDR, vitamin D receptor; TR, thyroid hormone
receptor; RAR, retinoic acid receptor; LXR, liver X receptor; PXR, pregnane X receptor; FXR, farnesoid X receptor; ER
n, everted repeat with
n
base-pair spacing; WY-14,643, [4-chloro-6-(2,3-xylidino)-2-pyrimidinylthio]acetic acid; tk, thymidine kinase promoter; LBD, ligand-binding do-
main; NCoR, nuclear receptor corepressor; VD3, 1␣,25-dihydroxycholecalciferol; RIF, rifampicin; RU486, mifepristone; ANDR, 3␣-androstenol;
CLOTR, clotrimazole; T3, tri-iodothyronine; COUP-TFI, chicken ovalbumin upstream promoter-transcription factor I; AF2, activation function-2;
trVD3, 1␣,25-dihydroxycholecalciferol; XREM, xenobiotic-responsive enhancer module; EE2, 17␣-ethynyl-3,17-estradiol.
Nuclear Receptor Cross-Talk on PB-Responsive CYP2B Enhancers
play considerable in vitro binding and activation of DR4-type
Steven Kliewer (GlaxoSmithKline, Research Triangle Park, NC).
sites (Mangelsdorf and Evans, 1995; Laffitte et al., 2000;
The mouse (mNR1) -tk-luc, human PBREM-tk-luc, and (hNR1) -tk-
Quack and Carlberg, 2000; Xie et al., 2000b). Therefore, it
luc plasmids have been described previously (Sueyoshi et al., 1999).
has been proposed that additional NRs could bind to PBREM
The human XREM-3A4-luc reporter containing the proximal 362
and modulate its activity (Waxman, 1999). This idea is in line
base pairs of
CYP3A4 gene promoter and the distal enhancer (Good-
with evidence that CYP2B mRNA induction is influenced by
win et al., 1999) was a kind gift from Dr. Chris Liddle (University ofSydney at Westmead Hospital, Westmead, Australia). The UAS -tk-
sex, steroid and thyroid hormones, sterol metabolites, and
luc (Janowski et al., 1996) and rat
CYP3A23[⫺1360/⫹82] reporters
retinoids, all of which are known NR ligands (Honkakoski
(Xie et al., 2000b) were donated by Dr. Ronald Evans (Salk Institute
and Negishi, 2000). Several inducers such as PB and pesti-
for Biological Studies, La Jolla, CA). Other luciferase reporter plas-
cides activate not only CAR (Sueyoshi et al., 1999) but also
mids for nuclear receptors were generated by inserting multiple
PXR, a receptor important for
CYP3A gene regulation (re-
copies of their cognate DNA sites into
BglII site of pGL3-Basic
viewed by Quattrochi and Guzelian, 2001). CAR and PXR
plasmid. All plasmids were purified with QIAGEN columns (Hilden,
recognize similar DNA motifs that range from DR2 to DR5
Germany) and verified by restriction mapping, functional testing
and everted repeat-6 (ER6), and both receptors are expressed
and, when necessary, by sequencing.
in the liver and intestine (Honkakoski and Negishi, 2000;
Expression Plasmids. The sources of expression vectors for
Quattrochi and Guzelian, 2001). Collectively, these data sug-
mRAR␣ and hRAR␣ (Zelent et al., 1989), cTR␣ (Harbers et al., 1996),
gest that other NRs might well affect the PBREM enhancer
hLXR (Teboul et al., 1995), mCOUP-TFI (NR2F1; Cooney et al.,
and influence
CYP2B gene regulation through cross-talk
1993), hPPAR␣ and mPPAR␣ (NR1C1; Sher et al., 1993), hFXR(Forman et al., 1995), hCAR and mCAR (Honkakoski et al., 1998;
Sueyoshi et al., 1999), hVDR (Quack and Carlberg, 2000) and hPXR
Surprisingly, there is very little information or systematic
and mPXR (Lehmann et al., 1998) have been described previously.
studies on PBREM binding or cross-talk with CAR by other
The expression plasmid for coactivator hTIF2 (Voegel et al., 1996)
NRs. Such studies are much needed for detailed understand-
was donated by Dr. Hinrich Gronemeyer (IGBMC, Illkirch, France).
ing of
CYP2B gene regulation, modulating factors, and spe-
GAL4-LBD Fusion Plasmids. The ligand binding domains (LBD)
cies differences. So far, we know that CAR can activate the
of mCAR (residues 118–358), hCAR (residues 108–348), mPXR (resi-
PXR-responsive ER6 and DR3 motifs in
CYP3A genes (Su-
dues 104–431), and hPXR (residues 107–434) were amplified with
Pfu
eyoshi et al., 1999; Moore et al., 2000; Xie et al., 2000b;
DNA polymerase from mouse and human liver RNAs and cloned into 5⬘
Smirlis et al., 2001). This is consistent with the report that
EcoRI and 3⬘
BamHI or
KpnI sites of CMX-GAL4 plasmid (Janowski et
PB can induce CYP3A mRNA in
PXR null mice (Xie et al.,
al., 1996) donated by Dr. Ronald Evans. GAL4-mCAR⌬8 plasmid coding
2000a). Recently, hPXR and mPXR were shown to bind to
for a truncated mCAR lacking eight amino acids at the C terminus
DR4 motifs and activate PBREM elements from various spe-
(Choi et al., 1997) was donated by Dr. David Moore (Baylor College ofMedicine, Houston, TX).
cies by 3- to 6-fold. This effect is roughly comparable with
Ligand Specificities of mCAR and hCAR. Ligand specificities of
that of CAR-mediated activation (Xie et al., 2000b; Goodwin
mCAR and hCAR were assessed for PBREM preference studies (Figs.
et al., 2001; Smirlis et al., 2001). None of these studies,
6-8) because CAR and PXR are reported to share some ligands (Moore
however, could address the preference of PBREM for CAR
et al., 2000) and to assess the effect of other NR ligands on mCAR and
and PXR. The data are also mostly based on in vitro DNA
hCAR activity. The ligand specificities were measured first by chemical-
binding assays with simple DR4 motifs (Sueyoshi et al., 1999;
dependent modulation of GAL4 fusion protein-driven reporter activity
Xie et al., 2000b; Smirlis et al., 2001) instead of functional
in HEK293 cells (Table 1) according to Honkakoski et al. (2001) and
assays with receptors and PBREM elements. Finally, there is
then by yeast two-hybrid assays as described below.
practically no data on the modulation of PBREM by other
Human and mouse CAR LBDs were inserted between
EcoRI and
NRs. Therefore, our aim was to gain more insight to the
BamHI sites in pGBKT7 plasmid. The NR interaction domains from
mouse and human PBREM function and its specificity for
mouse (residues 1988–2304) and human (residues 1972–2290) core-
CAR by evaluating the effects of several NRs on PBREM
pressor NCoR (Hu and Lazar, 1999) were cloned from liver RNAs
activity by functional and DNA binding assays. To help in
and inserted between
EcoRI and
BamHI sites in pGADT7 plasmid(Matchmaker GAL4 System 3, BD Clontech). All the manipulations
this effort, the ligand specificities of human and mouse CAR
were done essentially according to manufacturer's instructions. Ran-
were also defined.
dom yeast colonies selected on SD/Leu⫺/Trp⫺ plates were picked,amplified, and aliquots of cells were then treated with vehicle or testchemicals for 3.5 h before measurement of -galactosidase activities
Materials and Methods
and cell densities according to Nishikawa et al. (1999).
Chemicals. TCPOBOP was synthesized and purified according to
In Vitro Translation and Gel Shift Assays. NRs were produced
Honkakoski et al. (1996) to more than 98% purity as assessed by
in vitro by first transcribing linearized expression vectors with T7
1H-NMR spectra and elemental analysis (observed: N, 6.7%; C, 46.9%;
RNA polymerase and then translating these RNAs in vitro using
H, 2.0%; expected: N, 6.8%; C, 46.8%; H, 2.2%). Steroids were from
rabbit reticulocyte lysate as recommended by the supplier (Pro-
Steraloids, Inc. (Newport, RI) or Sigma-Aldrich Chemical Co. (St. Louis,
mega). Nuclear receptor heterodimers with RXR (approximately 10
MO). WY-14,643 was bought from ChemSyn, Inc. (Lenexa, KS). Other
ng of specific protein; equal protein amounts verified by a parallel
chemicals were at least analytical grade from Sigma, Fluka
translation in the presence of [35S]methionine) were incubated with
(Ronkonkoma, NY), or Calbiochem (La Jolla, CA).
ligand for 15 min at room temperature in a total volume of 20 l of
Reporter Plasmids. pCMV was purchased from BD Clontech
binding buffer [10 mM HEPES, pH 7.9, 1 mM dithiothreitol, 0.2
Inc. (Palo Alto, CA). The mPBREM-tk-luc reporter was constructed
g/l poly(dI-dC), and 5% glycerol], which was adjusted to 150 mM
by insertion of the PBREM element plus the thymidine kinase pro-
KCl. Approximately 1 ng of the 32P-labeled human
CYP2B6 or mouse
moter (tk) from the mouse PBREM-tk-CAT (Honkakoski et al., 1998)
Cyp2b10 DR4-type NR1 motif (50,000 cpm) was then added, and
into
BglII site of pGL3-Basic luciferase plasmid (Promega, Madison,
incubation was continued for 20 min. Protein-DNA complexes were
WI). The rat (rER6) -tk-luc reporter was constructed similarly from
resolved through 8% nondenaturing poly-acrylamide gels in 0.5⫻
(ER6) -tk-CAT plasmid (Lehmann et al., 1998) donated by Dr.
Tris/borate/EDTA (45 mM Tris, 45 mM boric acid, 1 mM EDTA, pH
Ma¨kinen et al.
8.3) and were quantified on a FLA3000 reader (Fuji, Tokyo, Japan)
0.5 M TCPOBOP, 10 M EE2, and 2 M CLOTR to varying
using Image Gauge software (Fuji).
degrees. With GAL4-hCAR, a reproducible partial deactiva-
Cell Culture and Nuclear Receptor Cotransfection. Mouse
tion (50–60%) by EE2 and about 2-fold activation by CLOTR
primary hepatocytes were isolated, transfected, and assayed as de-
was seen. Furthermore, the partial deactivation by EE2
scribed previously (Honkakoski and Negishi, 1998; Honkakoski et
could be overcome by addition of CLOTR and, to a lesser
al., 1998). HEK293 cells (American Type Culture Collection, Manas-sas, VA) were grown in phenol red-free Dulbecco's modified Eagle
extent, by 5-pregnanedione (Table 1). The known ligand
medium supplemented with 10% fetal bovine serum and 100 U/ml
profiles of mPXR and hPXR (Moore et al., 2000) were also
penicillin-100 g/ml streptomycin (Invitrogen, Gaithersburg, MD).
reproduced: both receptors were activated by CLOTR,
One day before transfection, the cells were seeded on 48-well plates
RU486, and 5-pregnanedione, but RIF activated only hPXR.
in medium containing delipidated serum (Sigma) to remove potential
Yeast two-hybrid assays supported the finding that ANDR
NR-activating substances. After an overnight incubation, the me-
and EE2 can deactivate mCAR and hCAR, respectively, be-
dium was changed and the cells were transfected using a calcium
cause they could dose-dependently increase association be-
phosphate method with pCMV (50 ng), various luciferase reporter
tween the CAR LBD and the NR interaction domain of the
plasmids (25 ng; 100 ng for XREM-3A4-luc and CYP3A23-luc), andvariable amounts of expression vectors for NRs (varied from zero to
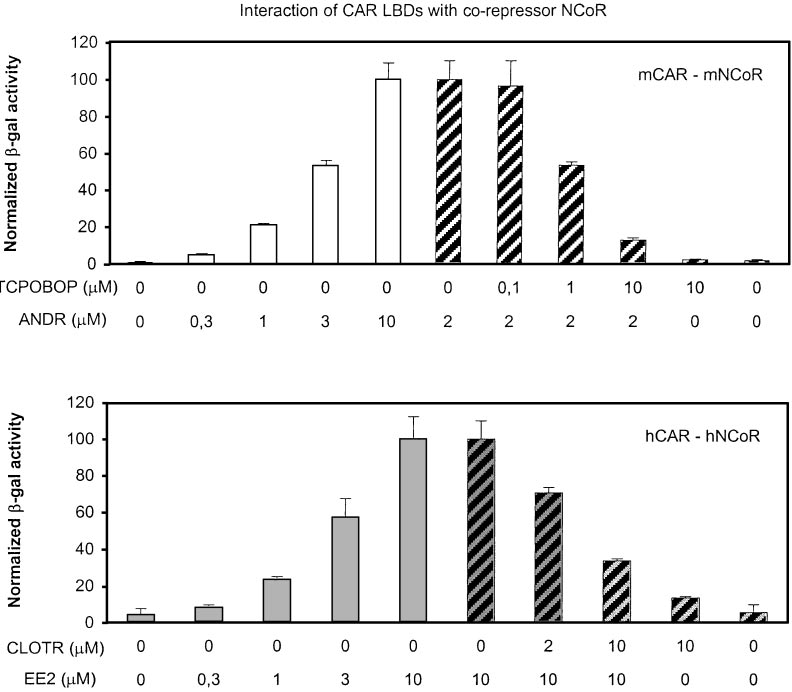
corepressor NCoR by 25- to 50-fold (Fig. 1, left, 䡺, u). This
250 ng). In activation and suppression experiments, the amount of
association could be reversed by TCPOBOP and CLOTR (Fig.
CAR expression plasmid that produced maximal activity from the
1, right, 3), whereas these activators themselves had little if
reporter plasmid was 12.5 ng, and other NRs were titrated from zero
any effect on the CAR LBD-NCoR interaction. These results
to 20-fold excess (250 ng) over CAR so as to reach the effect plateau.
obtained from two independent systems strongly suggest
In preference experiments, the total amount of NR expression vector
that the EE2 and CLOTR are true, reciprocally acting hCAR
was only 50 ng, much below levels that produced any unspecific
squelching (ⱖ200 ng). The balance of DNA was kept constant by
DNA Binding to NR1 Sites by NRs. PBREM elements
addition of empty expression vector.
After a 4-h transfection period, the medium was changed. The
are known to confer about 10-fold activation by CAR in
fresh medium additionally contained an established NR ligand/acti-
HEK293 cells and about 10-fold induction by TCPOBOP in
vator at concentration sufficient for maximal or near-maximal NR
primary hepatocytes (Honkakoski et al., 1998; Sueyoshi et
response: 20 M WY-14643 for h/mPPAR␣, 0.1 M 1␣,25-dihydroxy-
al., 1999). When organization of the mouse PBREM (5⬘ NR1-
cholecalciferol (VD3) for hVDR, 10 M rifampicin (RIF) for hPXR, 10
NFI-NR2 3⬘) was changed to NR1-NFI-NR1 or to NR2-NFI-
M mifepristone (RU486) for mPXR, 10 M 3␣-androstenol (ANDR)
NR2, the original and NR1-containing PBREM elements re-
or 0.5 M TCPOBOP for mCAR, 10 M 5-pregnanedione, 2 M
tained ⬎10-fold activation. PBREM containing NR2 motifs
clotrimazole (CLOTR), or 10 M 17␣-ethynyl-3,17-estradiol (EE2)
only conferred much lower ⬇3-fold activation by mCAR and
for hCAR (see Table 1), 10 M arotinoid acid for h/mRAR␣, 50 Mchenodeoxycholic acid for hFXR, 10 M 25OH-cholesterol for hLXR,
TCPOBOP inducibility (Fig. 2). This indicates that NR1 is
and 0.1 M tri-iodothyronine (T3) for cTR␣.
the stronger site for PBREM function.
Reporter Assays. Transfected HEK293 cells were cultured for
Gel shift assays were performed to compare the ability of
40 h, washed with PBS, and lysed. Luciferase and -galactosidase
NR/RXR␣ heterodimers to form complexes with NR1 sites
activities (Honkakoski et al., 2001) were determined from 20 l of
(Fig. 3). The human and mouse NR1 sites were similar in
lysates in 96-well plates using the Victor2 multiplate reader
their binding patterns. In the absence of specific activating
(PerkinElmer Wallac, Turku, Finland). All luciferase activities were
ligands, the ranking of complex formation was found to be
normalized to -galactosidase expression and expressed as mean ⫾standard deviation from three to four independent experiments.
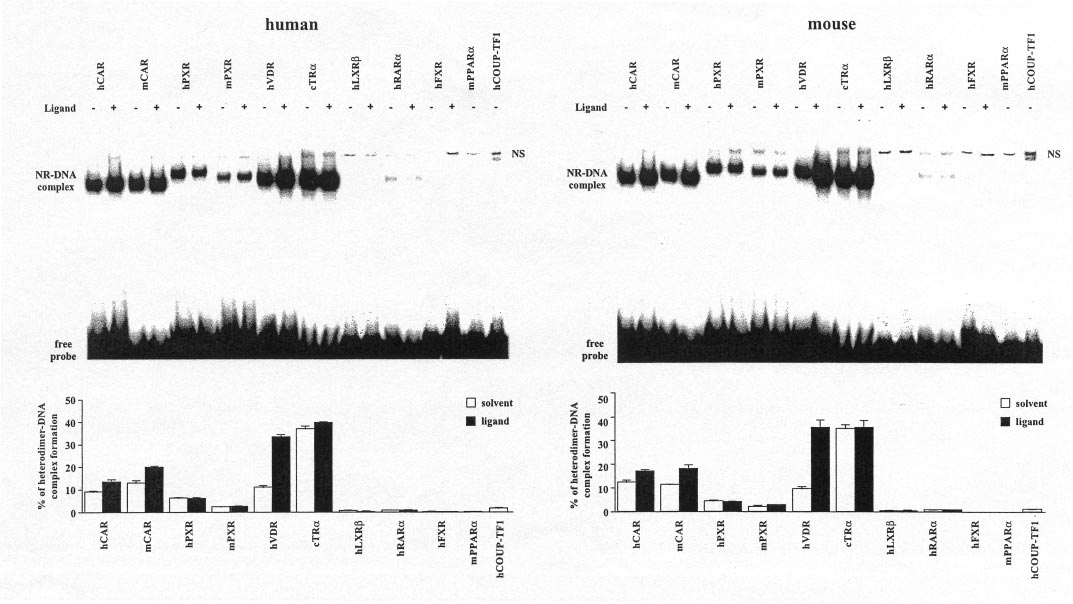
cTR␣ ⬎⬎ mCAR ⬎ hCAR ⬇ hVDR ⬎ hPXR ⬎ mPXR ⬇hCOUP-TFI ⬎ hRAR␣ ⬎ hLXR. Heterodimers of hFXR andhPPAR␣ showed no binding to NR1 sites but were demon-
strated to bind to consensus DR1-type motifs (data not
Ligand Specificities of mCAR and hCAR. With GAL4
shown). Addition of specific ligands enhanced complex forma-
fusion proteins in HEK293 cells (Table 1), we found that
tion only for mCAR, hCAR, and hVDR. However, the VD3-
GAL4-mCAR activity was suppressed by ANDR, as expected,
induced complex formation of hVDR was so strong that it
and that ANDR-suppressed activity could be reactivated by
practically equalled that of cTR␣. Thus, cTR␣ and hVDR
TABLE 1Ligand specificities of CAR and PXR with selected xenobioticsData are expressed as mean ⫾ S.D. (
n ⫽ 3) of fold activation of normalized luciferase activity. Other NR ligands (WY-14643, arotinoic acid, 25OH-cholesterol, VD3, and T3)were without any significant effect (ⱕ 15%) on hCAR or mCAR. Only chenodeoxycholic acid was a weak reactivator of ANDR-suppressed mCAR (2-fold). ANDR and EE2 didnot have significant effects on VDR-, PPAR␣-, or FXR-dependent activities.
12.52 ⫾ 1.20
a
1.70 ⫾ 0.15
a
0.70 ⫾ 0.11
a
1.41 ⫾ 0.12
a
8.18 ⫾ 0.23
a
0.41 ⫾ 0.05
a
1.97 ⫾ 0.02
a
1.50 ⫾ 0.13
a
1.83 ⫾ 0.23
a
1.73 ⫾ 0.06
a
2.26 ⫾ 0.23
a
3.21 ⫾ 0.58
aa
2.19 ⫾ 0.03
a
4.02 ⫾ 0.05
a
2.28 ⫾ 0.32
a
2.70 ⫾ 0.15
a
N.D., not done.
a Statistically different from vehicle (
p ⬍ 0.05).
Nuclear Receptor Cross-Talk on PB-Responsive CYP2B Enhancers
surpass both CAR and PXR isoforms in their ability to bind to
First, the maximal effect of NRs on simple DR4 motifs was
mouse and human NR1 sites.
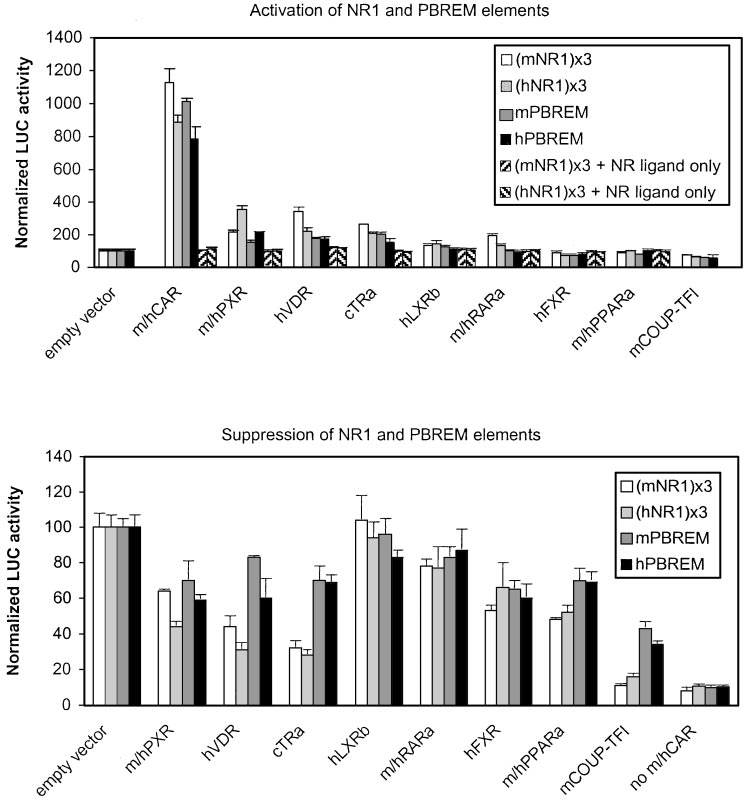
tested (Fig. 4, top, 䡺, u). Mouse (NR1) -tk-luc was activated,
Activation of NR1 Sites and PBREM Enhancers by
in descending order, by mCAR (11.2-fold) ⬎⬎ hVDR (3.4-
NRs. Binding to NR1 may indicate a potential influence on
fold) ⬎ cTR␣ (2.6-fold) ⬎ mPXR (2.0-fold) ⬇ mRAR␣ (1.9-
PBREM activity. To compare between NRs, increasing
fold). Addition of hLXR resulted in a slight 30% increase in
amounts of NR expression vectors were cotransfected with
activity, whereas COUP-TFI suppressed it by 25%. Human
NR1- or PBREM-driven reporter genes. The NRs were then
(NR1) -tk-luc was activated by hCAR (8.9-fold) ⬎⬎ hPXR
activated by established ligands, and the reporter activities
(3.5-fold) ⬎ hVDR (2.2-fold) ⬇ cTR␣ (2.1-fold). Human LXR
were measured. The results shown below are the maximal
and mRAR␣ increased and hFXR and COUP-TFI decreased
effects observed for each NR, usually at the same concentra-
the human NR1-driven activity slightly. Control experiments
tion as the optimal CAR concentration. The NRs themselves
with tk-luc plasmid lacking any enhancers established the
could ligand-dependently activate reporters driven by their
specificity of NR effects (data not shown). In addition, control
consensus response elements (data not shown), demonstrat-
experiments with activating NR ligands (Fig. 4, top, o, p)
ing that the constructs were functional.
showed that NR1-elements were not activated in the absence
Fig. 1. Ligand-dependent associa-
tion of mouse and human CAR LBD
with NR corepressor. Aliquots of
yeast cells transformed with GAL4-
mCAR LBD (top) or GAL4-hCAR
LBD (bottom) plus NR interaction
domain from NCoR plasmids were
treated for 3.5 h with vehicle or test
chemicals at indicated concentra-
tions (micromolar) before cell lysis
and -galactosidase assays as de-
scribed under Materials and Meth-
ods. For mCAR-mNCoR associa-
tion, the reporter activity with 10
M ANDR was set to 100 (top, 䡺).
In TCPOBOP displacement experi-ment (top, 3), the activity with 2
M ANDR was set to 100 (sameconcentration as in mammalianGAL4 assays in Table 1). ForhCAR-hNCoR association, the re-porter activity with 10 M EE2 wasset to 100 (bottom, u, 3). The datashown are mean ⫾ standard devia-tion from triplicate samples. Theexperiments were repeated inde-pendently two (mCAR) or threetimes (hCAR) with similar results.
Activities with either GAL4-CAR orNCoR plasmid alone were below de-tection limit.
Fig. 2. Activation of mutated mouse PBREM elements.
Reporter
PBREM sequence, NR1 sites only, NR2 sites only, or noenhancer (tk only) was cotransfected in the presence ofeither empty or CAR expression vector into HEK293 cells(䡺). Mouse hepatocytes were electroporated with the samereporters and treated with DMSO or 0.5 M TCPOBOP(f). Reporter activities from 3– 4 independent experimentswere measured as described under Materials and Meth-ods.
Ma¨kinen et al.
of NR expression vectors. In summary, almost all NRs capa-
20-fold excess over CAR) were used and effects at plateau
ble of NR1 binding in vitro were able to activate NR1-driven
only (typically 10-fold excess) are shown for clarity.
gene transcription to varying degrees, whereas COUP-TFI
Figure 4, bottom, indicates that mCAR-activated NR1-
inhibited it.
driven activity (Fig. 4, 䡺) was suppressed most efficiently by
When the same experiment was done with PBREM-driven
COUP-TFI (to 11% of control activity), followed by cTR␣
reporters (Fig. 4, top, f), a more restricted and attenuated
(32%) and hVDR (44%). Mouse PPAR␣, hFXR, and mPXR
response to NRs was noted. Mouse CAR was by far the
displayed a comparable 50 to 60% decrease, followed by
strongest activator of the mPBREM (10.1-fold), followed by
mRAR␣ and hLXR. Human CAR (Fig. 4, u) was suppressed,
ⱕ2-fold activation by cTR␣, hVDR, and mPXR. The human
in descending order, by COUP-TFI (to 16% of control activ-
PBREM was activated, in descending order, by hCAR (7.9-
ity) ⬎ cTR␣ ⬇ hVDR (about 30%) ⬎ hPXR ⬇ hPPAR␣ (about
fold) and ⱕ2.1-fold by hPXR, hVDR, and cTR␣. Human RAR␣
50%), followed by hFXR (65%). Again, hRAR␣ and hLXR
did not affect human PBREM, even though the NR1 element
had little or no effect. A less prominent suppression by the
was modestly but reproducibly activated. In addition, the
NRs was found on PBREM-driven reporters (Fig. 4, bottom,
extent of activation by other NRs was always less on PBREM
, f). Instead of the 50 to 90% decrease in activity that was
than on NR1 sites. For instance, the activation of NR1 by
observed on NR1 sites with COUP-TFI, cTR␣, hVDR, PXR, or
hVDR or PXR reached 28 and 40% of that by CAR, respec-
PPAR␣ isoforms, PBREM was inhibited by only 20 to 60% by
tively. On PBREM, hVDR and PXR reached only 18 and 27%
the same receptors. There was also a tendency for human
of CAR-dependent activity. PBREM seems to be activated
NR1 and PBREM to be inhibited more than corresponding
preferentially by CAR and then by similar efficiency (ⱕ2-
mouse elements by NR1-binding PXR isoforms, hVDR, and
fold) by cTR␣, hVDR, and PXR. Mouse PXR was a poorer
cTR␣. In line with activation results, mPXR was a weaker
activator of both NR1 and PBREM elements than hPXR.
suppressor than hPXR. In the context of PBREM, hFXR, and
PPAR␣ isoforms suppressed CAR as efficiently as PXR iso-
PBREM Enhancers by NRs. Because NRs may influence
forms hVDR and cTR␣, which bind to and inhibit NR1 more
PBREM function by competing for DNA binding sites or for
common NR coregulators, NRs were cotransfected in the
Suppressive Effects of NRs Occurring through CAR
presence of CAR and the maximal NR-mediated suppression
LBD. Expression vectors for GAL4-m/hCAR and UAS -tk-luc
of CAR-dependent NR1- or PBREM-driven activities were
reporter were used in the above suppression assay to deter-
analyzed. Increasing amounts of NR expression vectors (0- to
mine whether suppression could be attributed to competition
Fig. 3. RXR␣-Heterodimer complex formation of various NRs on the human (left) and mouse (right) NR1 sites. Gel shift experiments were performed
with in vitro translated heterodimers of equal amounts of the indicated NRs with RXR␣ that were preincubated at room temperature with saturating
concentrations of activators (f) [5-pregnanedione (hCAR), TCPOBOP (mCAR), RIF (hPXR), RU486 (mPXR), VD3 (hVDR), T3 (cTR␣), 25OH-
cholesterol (hLXR), all-trans-retinoic acid (hRAR␣), chenodeoxycholic acid (hFXR)] or solvent (䡺) and the 32P-labeled hNR1 or mNR1 site.
Protein-DNA complexes were separated from the free probe through 8% nondenaturing polyacrylamide gels. Representative experiments are shown.
The amount of heterodimer-DNA complexes in relation to free probe was quantified by bioimaging. Columns and bars indicate mean and S.D.,
respectively, from three experiments. NS, nonspecific complex.
Nuclear Receptor Cross-Talk on PB-Responsive CYP2B Enhancers
for factors associated with the CAR LBD. This approach
effect of NRs for PBREM enhancers. Furthermore, the activa-
would eliminate any competition at the level of DNA binding,
tion and suppression experiments yielded information on only
which was prominent for cTR␣, hVDR, and PXR isoforms.
the maximal effect by an NR, not on the preference of PBREM
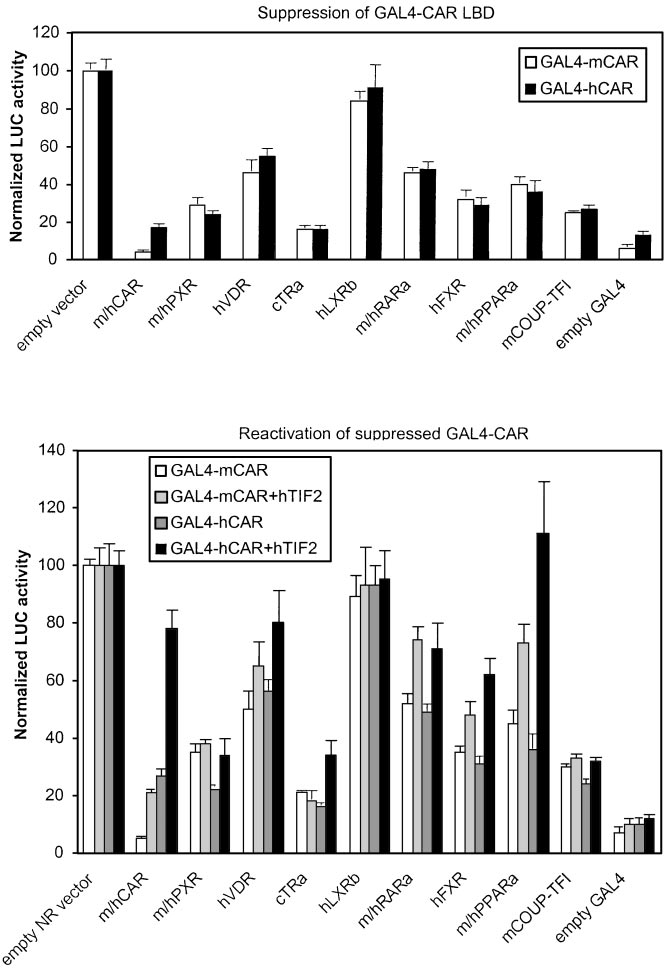
Figure 5, top, indicates that with both GAL4-mCAR and
for a particular NR. Therefore, detailed titrations with selected
GAL4-hCAR as activators, the strongest suppressors were
NR1-binding NRs were performed. The transfected cells were
full-length CAR and cTR␣ (⬍20% of control activity), fol-
treated with CAR-deactivating chemical and an activator spe-
lowed by COUP-TFI ⬇ PXR isoforms ⬇ hFXR (25–30%),
cific for the competing NR. We selected ANDR and EE2 for
whereas hVDR, RAR␣, and PPAR␣ isoforms were weaker
mCAR and hCAR, respectively, because ANDR can completely
suppressors (35–55%). No suppression was seen with GAL4-
deactivate mCAR (Forman et al., 1998; Sueyoshi et al., 1999),
mCAR⌬8 that lacks the AF2 core sequence (data not shown),
and EE2 is a partial deactivator of hCAR (Table 1, Fig. 1). In
indicating that NR-mediated suppression of CAR depends on
contrast, ANDR and EE2 had either a slight positive effect on
the presence of intact AF2 domain. Therefore, suppression of
PXR isoforms (Table 1) or did not affect other NRs at all (see
CAR LBD probably reflects competition for NR coactivators.
below). These "reciprocal" effects on NR activity allowed us to
Indeed, cotransfection of TIF2 vector in this suppression
better assess the functional preference of PBREM.
assay (Fig. 5, bottom) resulted in partial restoration of
It should be noted that in preference studies, much lower
mCAR-dependent and especially hCAR-dependent reporter
total amounts of NRs than in suppression assays (Fig. 4,
activity. Differences in the extent of suppression and resto-
bottom) were used (50 versus 250 ng), and therefore unspe-
ration of reporter activity further imply that NRs may have
cific squelching effects are not likely. In activation experi-
different affinities for various NR coactivators. In summary,
ment titrations (data not shown), we did not see any signif-
NR1-binding cTR␣ and hVDR showed a remarkable differ-
icant self-suppression by increasing amounts of CAR, PXR,
ence in their ability to suppress CAR LBD. PPAR␣ isoforms
or any other NR plasmid. This would happen if NR coregu-
and hFXR that do not bind to NR1 sites could inhibit PBREM
lators were a limiting factor and result in squelching. This
through an AF2- and coactivator-dependent mechanism.
did not seem to be the case.
Preference of NRs for PBREM Enhancers. As shown
Figure 6, top, shows that in the presence of saturating
above, DNA binding studies were not sufficient to assess the
amounts of mPXR only, mPBREM was activated 2-fold by
Fig. 4. Activation and suppression of
mouse and human NR1- and PBREM-
driven reporter genes by various NRs. Ac-
tivation by NRs (top) was assessed by
cotransfection into HEK293 cells of in-
creasing amounts of indicated NR expres-
sion vectors (0 –250 ng) and reporter
genes (25 ng) driven by mNR1 (䡺), hNR1
(u), mPBREM ( ), or hPBREM (f) ele-
ments and addition of NR-specific li-
gands. o, p, ligand controls (empty vector
⫹ activating NR ligand). Representativeresults at optimal 12.5 ng of NR expres-sion vectors (37.5 ng for RAR␣) areshown, with columns and bars denotingmean and S.D., respectively. Suppressionby NRs (bottom) was assessed by cotrans-fection of increasing amounts of indicatedNR expression vectors (0 –250 ng) to-gether with saturating amount of mouseor human CAR expression vector (12.5ng) and reporter genes (25 ng) driven bymNR1 (䡺), hNR1 (u), mPBREM ( ), orhPBREM
HEK293 cells were treated with NR-spe-cific ligands. Activity with empty vectorwas set to 100. Representative results atmaximal effect (125 ng of NR expressionvectors) are shown, with columns andbars denoting mean and S.D., respec-tively. No CAR, basal reporter activity inthe presence of empty expression vectorsubstituted for both CAR and the compet-ing NRs.
Ma¨kinen et al.
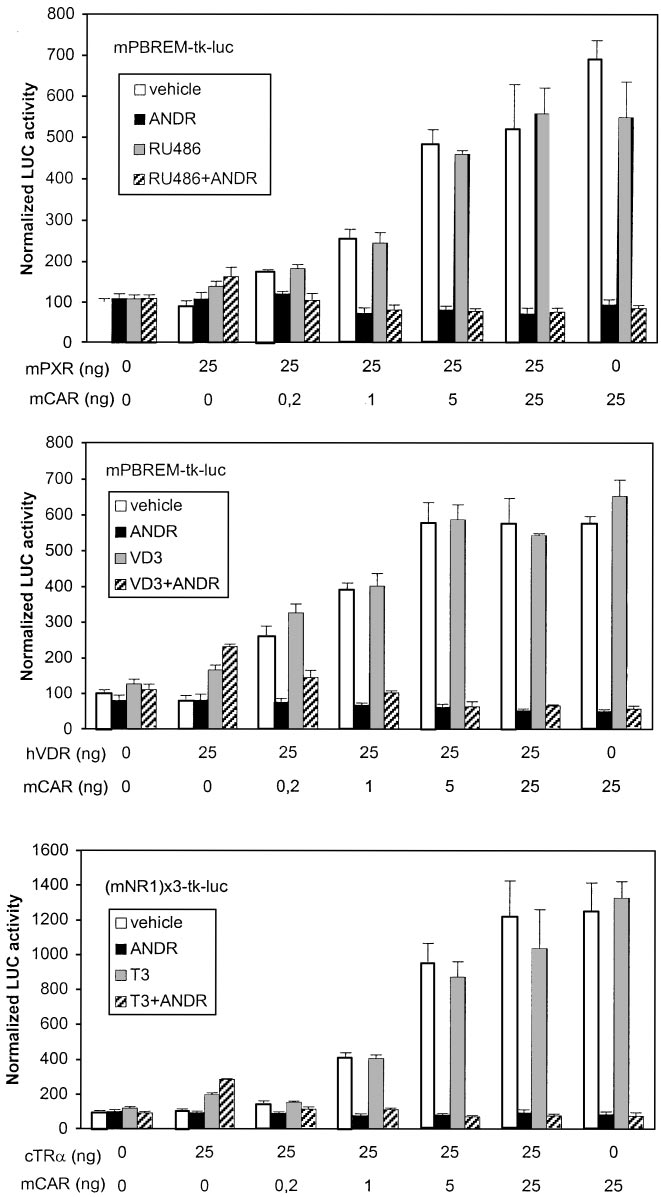
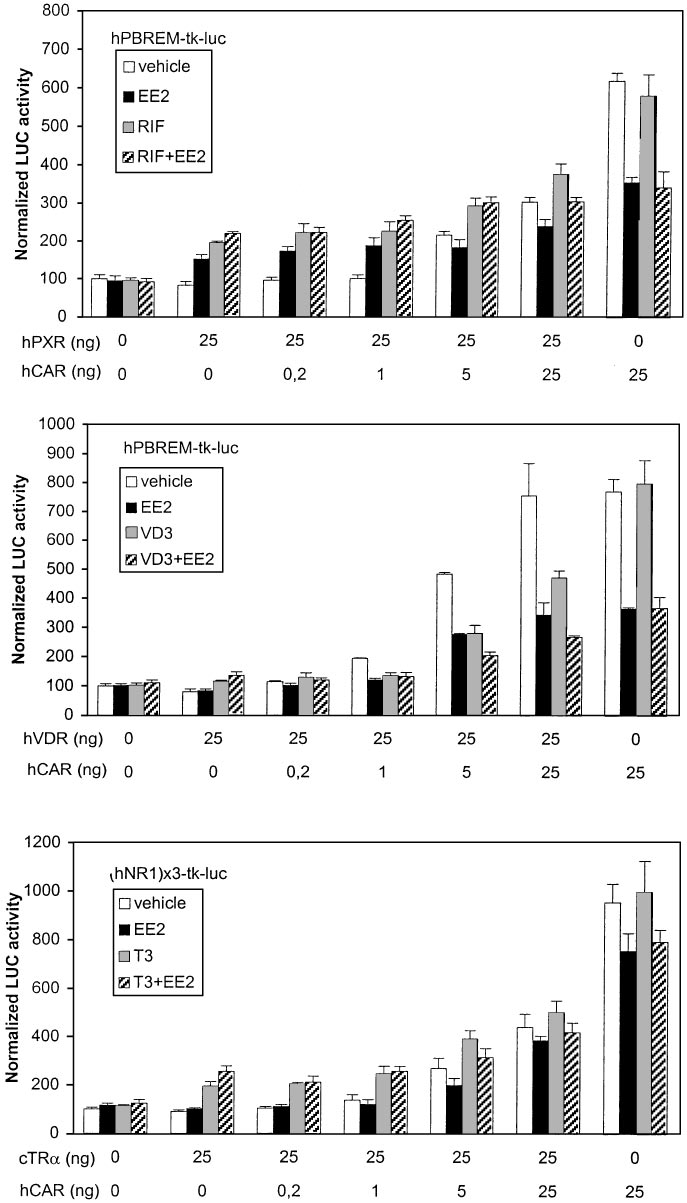
RU486, regardless of the presence of ANDR, as expected from
ratios of mCAR to cTR␣, the combined T3⫹ANDR treatment
data in Table 1. In the presence of mCAR only, mPBREM was
decreased activities to control levels, suggesting a strong mCAR
activated 7-fold, which was completely abolished by ANDR.
dominance over cTR␣.
Already at 1:25 ratio of mCAR to mPXR expression vectors, the
Human PBREM was activated 1.8-fold by EE2 and 2.5-fold
combined RU486⫹ANDR treatment suppressed the mPBREM-
by RIF in the presence of hPXR only. As predicted, hPBREM
driven activity to control levels, suggesting that mCAR clearly
activity was decreased to 40% by EE2 but not affected by RIF
dominates mPXR on PBREM. Mouse PBREM was activated
in the presence of hCAR only (Fig. 7, top). Compared with
2-fold by 0.1 M VD3 in the presence of hVDR (Fig. 6, middle).
results with mCAR-to-mPXR titration on mPBREM, hPXR
When the ratio of mCAR to hVDR was increased stepwise, the
predominated strikingly over hCAR at a 1:25 ratio, showed
combined VD3⫹ANDR treatment gave slightly higher LUC
substantial activity at a 5:25 ratio; hCAR predominated only
activities than ANDR alone up to 1:25 ratio, suggesting that
at a 25:25 ratio. Cotransfection of hVDR (Fig. 7, middle) that
hVDR binds to mPBREM slightly more avidly than mPXR.
combined VD3⫹EE2 treatment reduced the activity below
However, the hVDR-mediated suppression at 5:25 ratio (about
those elicited by VD3 alone beginning at hCAR-to-hVDR
15%) was less than for mPXR. Both these results are well in line
ratio of 5:25. In contrast to mPBREM, hPBREM activity was
with DNA binding and GAL4-mCAR suppression studies (Figs.
substantially reduced (to 60%) by VD3, matching the similar
3 and 4, bottom). Because of very modest activation of mP-
difference seen in suppression assay in Fig. 4. On human
BREM by cTR␣ (see Fig. 3), the experiment was done with
NR1 sites, cTR␣ was the dominant receptor up to 5:25 ratio
mNR1 reporter (Fig. 6, bottom). Already at 1:25 and greater
of hCAR to cTR␣, above which the combined T3⫹EE2 treat-
Fig. 5. Suppression of GAL4-CAR-activated reporter
activities by various NRs. Top, suppression by NRs
was assessed by cotransfection of increasing amounts
of indicated NR expression vectors (0 –250 ng) to-
gether with saturating amount of mouse (䡺) or hu-
man (f) GAL4-CAR LBD expression vector (12.5 ng)
and UAS -tk-luc reporter (25 ng) into HEK293 cells,
followed by addition of NR-specific ligands. Reporteractivity with empty vector was set to 100. Represen-tative results at maximal effect (125 ng of NR expres-sion vectors, 10-fold excess over CAR) are shown,with columns and bars denoting mean and S.D., re-spectively. GAL4 only, basal activity in the absence ofany LBD in the construct. Bottom, reactivation ofsuppressed GAL4-mCAR- or GAL4-hCAR-dependentactivity was performed as above with cotransfection(500 ng) of empty expression vector (䡺, ) or hTIF2plasmid ( , f).
Nuclear Receptor Cross-Talk on PB-Responsive CYP2B Enhancers
ment began to decrease the activity (Fig. 7, lower). These
present. XREM activity driven by hCAR only was again
results show that human PBREM was less selective for CAR
decreased 50% by EE2 but not affected by RIF. Titration with
than mouse PBREM.
increasing amounts of hPXR vector indicated that the de-
creasing effect of EE2 on XREM-driven activity was lost only
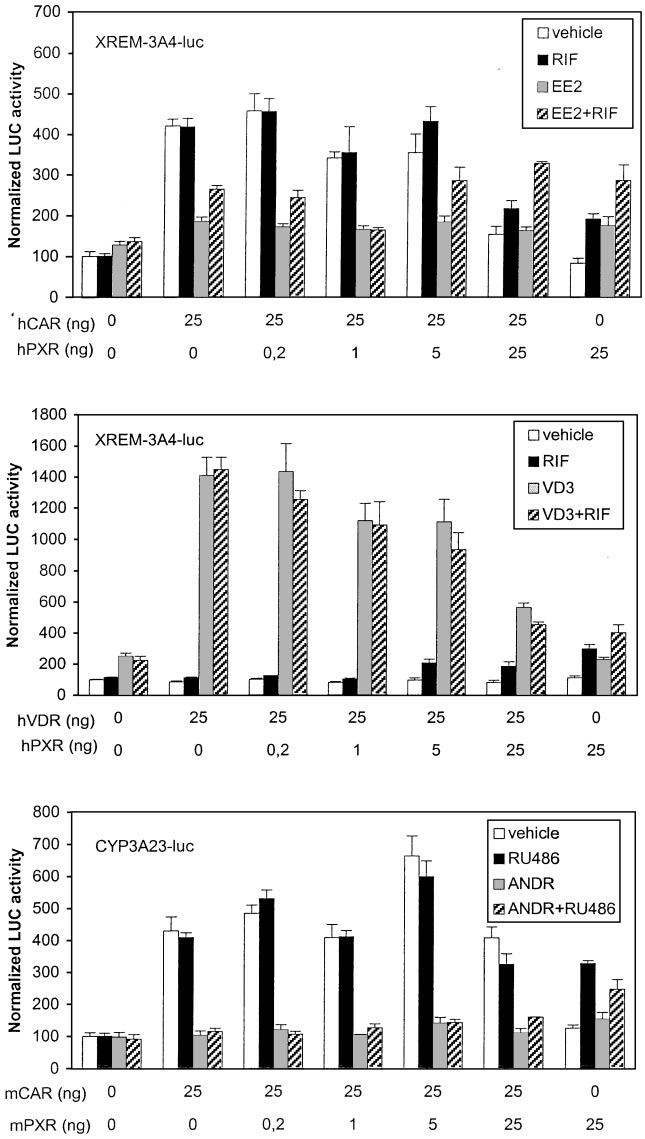
CYP3A23-luc reporters were used in similar preference stud-
at 25:25 ratio of hPXR to hCAR. This suggests that hCAR has
ies with hCAR, mCAR, and hVDR. Figure 8, top, shows that
considerable affinity to XREM motifs ER6 and/or DR3. Ex-
the human XREM enhancer was activated 2- to 3-fold by
periments with (rER6) -tk-luc reporter proved that at least
EE2, RIF, and their combination when hPXR only was
ER6-driven activity could be enhanced comparably by hCAR
Fig. 6. Dominance of mCAR over other NRs on mP-
BREM. Activation of mPBREM- or mNR1-driven re-
porter genes (25 ng) was assessed by cotransfection of
indicated amounts of NR expression vectors (0 –25 ng)
into HEK293 cells, with balance of DNA kept constant
by addition of empty expression vector. The trans-
fected cells were treated with vehicle (䡺), mCAR-
specific deactivator ANDR (f), competitor NR-specific
activating ligand (u), or their combination (o). Activ-
ity with empty vector plus vehicle only was set to 100.
Columns and bars denote mean and S.D., respec-
tively, from three independent experiments.
Ma¨kinen et al.
(6-fold) and hPXR (4-fold) (data not shown). Transfection of
that (rER6) -tk-luc and CYP3A23-luc reporters were induced
hVDR and VD3 treatment increased XREM-driven activity
5- and 20-fold, respectively, by ligand-activated hVDR (data
very strongly (Fig. 8, middle). Transfection of equal amounts
not shown). Figure 8, bottom, shows that mCAR and mPXR
of hPXR and hVDR resulted in more than 60 and 40% sup-
activated the CYP3A23-luc reporter over 4- and 3-fold, re-
pression of hVDR- and hPXR-dependent activities, respec-
spectively. Mouse CAR has substantial activity over mPXR,
tively. This suggested similar competition between hVDR
because the combined RU486⫹ANDR treatment began to
and hPXR for the XREM binding sites but higher activation
increase reporter activity above ANDR levels only at 25:25
potential by hVDR. This notion was supported by the finding
ratio of mPXR to mCAR. Similar mCAR dominance over
Fig. 7. Dominance of hCAR over other NRs on
hPBREM. Activation of hPBREM- or hNR1-driven
reporter genes (25 ng) was assessed as in Fig. 6.
The transfected cells were treated with vehicle
(䡺), hCAR-specific partial deactivator EE2 (f),
competitor NR-specific activating ligand (u) or
their combination (o).
Nuclear Receptor Cross-Talk on PB-Responsive CYP2B Enhancers
mPXR were seen with (rER6) -tk-luc reporter (data not
of CYP2B gene regulation, mechanisms, and species differ-
shown). Our results indicated that, in contrast to PBREM
ences therein. Our studies were aimed at resolving the func-
elements more selective for CAR, the CYP3A enhancers are
tional interplay between several NRs expressed in the liver
very responsive to hVDR, CAR, and PXR isoforms.
and the mouse and human PBREM elements. To help in thistask, CAR ligand binding specificities had to be defined in
more detail as well.
The preference of PBREM for various NRs is not known
Ligand Specificities of mCAR and hCAR. The known
although many NRs can bind to DR4-type motifs contained in
ligand profiles of mCAR and PXR isoforms (Forman et al.,
PBREM. Thus, assessment of NRs with respect to their
1998; Lehmann et al., 1998; Sueyoshi et al., 1999; Moore et
PBREM-modulating activity is important for understanding
al., 2000) were well reproduced in our GAL4 fusion protein
Fig. 8. Dominance of hPXR and mPXR over other NRs
on CYP3A enhancers. Activation of XREM-3A4- or
CYP3A23-driven reporter genes (100 ng) was assessed
as in Fig. 6. The transfected cells were treated with
vehicle (䡺), hPXR-specific RIF or mPXR-specific RU486
(f), competitor NR-specific ligand (u), or their combina-
tion (o).
Ma¨kinen et al.
assays. With respect to hCAR, we confirmed that 5-preg-
ple DR4 motifs from the repressive effects, as shown by
nanedione was a modest activator, TCPOBOP had no ef-
diminished suppression by, for example, cTR␣, hVDR, PXR,
fect, and ANDR was a weak deactivator, as shown earlier
and PPAR␣ isoforms. Only COUP-TFI, a well-known sup-
by Moore et al. (2000). Intriguingly, EE2 proved to be an
pressor (Cooney et al., 1993), could bring the CAR-dependent
activator of mCAR but a partial deactivator of hCAR.
PBREM activity below 50%. We observed that hPBREM is
Because HEK293 cells do not express estrogen receptors
notably less selective for CAR and more prone to NR-medi-
(Kahlert et al., 2000), EE2 cannot inhibit hCAR activity
ated suppression than mPBREM. In hPBREM, the NFI site
via estrogen receptor-dependent squelching. EE2 was
seems to be mutated (Sueyoshi et al., 1999); therefore, NFI
found to strongly promote the interaction between hCAR
might play a role in the high selectivity of mPBREM. Kim et
LBD and a NR corepressor, lending strong support for
al. (2001) have recently shown that NFI and CAR can bind
direct inhibitory action of EE2 on hCAR. In contrast to the
simultaneously to rat PBREM in vitro and that NFI coex-
report by Moore et al. (2000), we did not detect any sup-
pression may enhance trans-activation by CAR. This attrac-
pression by CLOTR of hCAR activity. Instead, CLOTR
tive mechanism cannot yet explain the selectivity of PBREM
activated GAL4-hCAR on its own and could also overcome
for CAR because CAR and NFI bound independently of each
the inhibition by EE2. This finding was also supported by
other, at least in vitro, and other NRs could potentially sub-
our yeast two-hybrid experiments. Moore et al. (2000) re-
stitute for CAR. It may be possible that specific cofactors,
ported decreases by CLOTR in hCAR activity in CV-1 cells
lacking from in vitro studies, mediate the interaction be-
and in in vitro association between hCAR LBD and coac-
tween NFI and CAR. Other possibilities include co-operation
tivator SRC-1 with a FRET-based assay. Perhaps cell- and
between CAR-bound NR1 and NR2 sites that cannot be re-
assay-specific differences may explain these differences.
produced on multimeric NR1 sites. This option is consistent
For instance, the decrease by ANDR of mCAR–SRC1 in-
with the earlier report that mutation of any NR half-site in
teraction that was seen in a GST pulldown assay (Forman
mPBREM reduced the PB inducibility to a similar extent
et al., 1998) could not be reproduced by Moore et al. (2000).
(Honkakoski et al., 1998). Further studies into these hypoth-
In our view, deactivators may be studied best with core-
eses are warranted.
pressor association assays.
Preference of PBREM and XREM for NRs. The NR
NR1 Binding and Activation Specificity. Although
preference studies indicated that mCAR predominates on
NR1 sites alone confer CAR responsiveness (Sueyoshi et al.,
mPBREM over weaker effectors such as mPXR. Therefore,
1999), the presence of both NR1 and NR2 sites in the natural
only weak activation by pure NR ligands of Cyp2b10 gene
PBREM enhancer seems crucial for optimal activation
might be expected in vivo. Indeed, hepatocytes transfected
(Honkakoski et al., 1998; Goodwin et al., 2001). Despite pre-
with a PBREM construct showed only 2-fold activation after
vious observations that mouse CAR/RXR␣ heterodimer binds
PCN treatment (Xie et al., 2000b; Smirlis et al., 2001); he-
to NR1 and NR2 sites with equal efficiency in vitro (Tzameli
patic CYP2B10 was induced 37-fold by PB but only 7-fold by
et al., 2000), the present functional studies indicated that
PCN (Pellinen et al., 1994); CYP2B10 mRNA induction was
NR1 site is the stronger of these DR4 motifs. Paquet et al.
not affected either by thyroid hormone or by retinoic acid in
(2000) have also suggested that NR1 and NR2 in rat CYP2B2
mouse hepatocytes (Honkakoski and Negishi, 1998); T3 does
gene are not identical. Among many NRs capable of DR4
not seem to affect PBREM or its associated factors in rats
binding, only hPXR and mPXR have been reported to bind to
(Ganem et al., 1999). The identity of 5⬘-flanking nucleotides
the NR1 site with affinity similar to CAR (Xie et al., 2000b;
in DR4 motifs is important for TR-mediated activation (Har-
Goodwin et al., 2001; Smirlis et al., 2001). Here, many other
bers et al., 1996; Zhang and Lazar, 2000) and this property
NRs were assessed through in vitro translation and NR1
may explain the discrepancy between the strong binding and
probe binding under optimized conditions. Human VDR and
inefficient function by TR␣ on PBREM. Although TR iso-
cTR␣ bound to NR1 with greater efficiency than CAR, which
forms are expressed in liver (Zhang and Lazar, 2000), and TR
in turn displayed better binding than PXR isoforms. If the
can inhibit CAR LBD, the levels of TR relative to CAR may be
binding efficiency to NR1 were the sole determinant of
too low for significant suppression via competition for NR
PBREM activation, then one would predict that cTR␣ and
coregulators. On the other hand, hPBREM seems to allow
hVDR would be strong activators of PBREM. Clearly, this
some hVDR and especially hPXR interactions. This probably
was not the case. On simple NR1 sites, activation by CAR
explains why RIF, a specific hPXR ligand, can efficiently
greatly surpassed that of cTR␣, VDR, or PXR, which showed
induce CYP2B6 mRNA in human hepatocytes (e.g., Goodwin
a maximal 2- to 3.5-fold activation. On natural PBREM ele-
et al., 2001). To our knowledge, there are no data available on
ments, these three receptors were even less efficient. This is
the response of CYP2B genes to VDR ligands or VDR status.
in contrast with the results of Smirlis et al. (2001) and Xie et
The CYP3A4 and CYP3A23 enhancers, in contrast, re-
al. (2000b), who found similar or 40% smaller activation of
spond not only to PXR but also to CAR and VDR. These
rodent PBREMs by mPXR or hPXR than by mCAR, respec-
experiments now give, for the first time, a mechanistic ex-
tively. However, they found ⬃2-fold activation by PCN of
planation of the strong inducibility of Cyp3a genes by the
PBREM in hepatocytes, which is similar to the 2- to 2.5-fold
mCAR ligand TCPOBOP (e.g., Smith et al., 1993), to the
activation by RU486 seen in HEK293 cells.
induction of CYP3A4 mRNA by VD3 in Caco-2 cells
NR Cross-Talk Is Attenuated on PBREM Elements.
(Schmiedlin-Ren et al., 1997), and suggest why CYP3A and
The activation potential of NRs was significantly weaker on
CYP2B genes tend to be coregulated in humans. Our results
PBREM enhancers compared with NR1 sites. This suggests
are in contrast with Moore et al. (2000), who found that hPXR
that NR interaction with DR4 motifs imbedded in PBREM is
predominated hCAR on the XREM enhancer. Their conclu-
restricted, resulting in increased specificity for CAR. Fur-
sion was based on the repressive effect of CLOTR on hCAR,
thermore, PBREM enhancers are more ‘insulated' than sim-
a finding that we could not reproduce with either full-length
Nuclear Receptor Cross-Talk on PB-Responsive CYP2B Enhancers
TABLE 2Summary of NR effects on CAR signaling
Activation of NR1/
Suppression of NR1/
Predicted Relevance of CAR
Cross-Talk in Vivo
⫹⫹/⫹⫹ b
⫹⫹⫹⫹ Human, ⫹⫹ Mouse
⫹⫹⫹/⫹ b
⫹⫹ Human, 0 Mouse
⫹ Human, 0 Mouse
⫹ Human, ⫹⫹ Mouse
⫹⫹⫹⫹, Strong effect; ⫹⫹⫹, significant effect; ⫹⫹, moderate effect; ⫹, weak effect; 0, no effect; N.A., not applicable.
a COUP-TFI represses NR1 and PBREM.
b PXR, VDR, and COUP-TFI suppress hPBREM more than mPBREM.
c Hepatic expression of hPPAR␣ is significantly lower than that of mPPAR␣.
hCAR, GAL4 fusion plasmids, or with yeast two-hybrid as-
T3 suppresses CYP2B mRNA expression in rats but does not
affect PBREM (Ganem et al., 1999). Moreover, these experi-
Interference of CAR Signaling without Significant
ments would require truly monospecific NR ligands to avoid,
NR1 Binding. PPAR␣ and FXR that bind poorly if at all to
for example, interference with glucocorticoid signaling that is
NR1 sites can still significantly inhibit CAR-mediated sig-
essential for CYP2B regulation (Honkakoski and Negishi,
naling. This suppression seems to be caused by reversible
2000) that plagues the mPXR ligands PCN and RU486, and
competition for coactivators. Recent experiments with NR
the cross-activation of NRs such as FXR and PXR by bile
null mice suggest that the interference of CAR function,
acids. Given the lower selectivity of human PBREM and
detected here by cotransfection assays, may have physiolog-
XREM for NRs, human hepatocytes would probably be the
ical relevance in the liver where all these NRs are predomi-
best system in which to run the experiments. Most impor-
nantly expressed. For example, the lack of PPAR␣ greatly
tantly, tissue or hepatocytes from NR null mice would be
enhances the mitogenic effects of TCPOBOP (Columbano et
most valuable in confirming the observed NR interferences.
al., 2001) that are mediated by CAR (Wei et al., 2000). Be-
In conclusion, our results indicate that binding of an NR to
cause CAR/RXR␣ heterodimer binding is important for both
NR1 sites does not correlate with its functional effects in the
basal and inducible CYP2B gene expression (Wan et al.,
context of PBREM. The use of simple NR motifs for binding
2000; Wei et al., 2000), it is possible that PPAR␣ suppresses
and trans-activation assays may not reveal actual function of
CAR and exerts an effect on PBREM. The activation of
an NR on natural DNA elements. Mouse PBREM was found
CYP3A and CYP2B gene expression in the absence of FXR
to be more selective for CAR than human PBREM, which is
(Schuetz et al., 2001) is difficult to interpret similarly be-
also activated by PXR, VDR, and TR␣. In contrast to PBREM
cause bile acids that accumulate in FXR null mice are also
elements, CYP3A enhancers were highly responsive to VDR,
activators for PXR (e.g., Schuetz et al., 2001) and possibly
CAR, and PXR. PPAR␣ and FXR may use mechanisms de-
weak ligands for mCAR as well (see Table 1). An FXR-specific
pendent on coactivators to interfere with CAR signaling.
chemical probe should help resolve this question and furtherelucidate interactions between NRs and P450 gene expres-
We thank Drs. Pierre Chambon (IGBMC, Illkirch, France), Ronald
Finally, we have attempted to classify NRs based on the
Evans, Frank Gonzalez (NCI, Bethesda, MD), Hinrich Gronemeyer,
observed DNA binding and PBREM-suppressive effects (Ta-
Steven Kliewer, David Mangelsdorf, (University of Texas Southwest-
ble 2). This data combined with approximate hepatic levels of
ern Medical Center, Dallas, TX), Masahiko Negishi and Cary Wein-
mouse NRs reported in the literature (e.g., Wan et al., 2000;
berger (NIEHS, Research Triangle Park, NC), Ming-Jer Tsai (Baylor
Xie et al., 2000a; Zhang and Lazar, 2000) may allow us to
College of Medicine, Houston, TX), Bjo¨rn Vennstro¨m (Karolinska
predict the relevance of NR cross-talk with CAR signaling. As
Institute, Stockholm, Sweden), Steven Kliewer, Chris Liddle, David
positive signs of this predictability, the moderate or weak
Moore, for plasmids, and Kaarina Pitka¨nen for technical assistance.
efficiency of mPXR, TR␣, and RAR␣ for PBREM activation is
indeed reflected in some in vivo studies (Pellinen et al., 1994;
Choi HS, Chung M, Tzameli I, Simha D, Lee YK, Seol W, and Moore DD (1997)
Honkakoski and Negishi, 1998; Ganem et al., 1999). Simi-
Differential trans-activation by two isoforms of the orphan nuclear hormone re-
larly, PPAR␣ seems to suppress CAR activity in vivo (Colum-
ceptor CAR. J Biol Chem 272:23565–23571.
Columbano A, Ledda-Columbano GM, Pibiri M, Concas D, Reddy JK, and Rao MS
bano et al., 2001).
(2001) Peroxisome proliferator-activated receptor-␣(⫺/⫺) mice show enhanced he-
Collectively, our studies generate hypotheses for further in
patocyte proliferation in response to the hepatomitogen 1,4-bis[2-(3,5-dichloro-
pyridyloxy)]benzene, a ligand of constitutive androstane receptor. Hepatology 34:
vivo experiments that must, however, be carefully controlled
to rule out any nonspecific effects occurring outside PBREM.
Cooney AJ, Tsai SY, O'Malley BW, and Tsai M-J (1993) Chicken ovalbumin up-
stream promoter transcription factor (COUP-TF) dimers bind to different GGTCA
As examples of the associated problems, CYP3A and CYP2B
response elements, allowing COUP-TF to repress hormonal induction of the vita-
genes contain NR binding sites also outside of XREM and
min D3, thyroid hormone and retinoic acid receptors. Mol Cell Biol 12:4153– 4163.
PBREM. For examples, the CYP3A promoter contains DR1
Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka
LT, McMorris T, Kozak CA, et al. (1995) Identification of a nuclear receptor that
sites that are crucial for CYP3A basal activity (Quattrochi
is activated by farnesol metabolites. Cell 81:687– 693.
and Guzelian, 2001) but are also targets for COUP-TFI,
Forman BM, Tzameli I, Choi HS, Chen J, Simha D, Seol W, Evans RM, and Moore
DD (1998) Androstane metabolites bind to and deactivate the nuclear receptor
hepatocyte nuclear factor-4, and perhaps other NRs as well.
CAR-. Nature (Lond) 395:612– 615.
Ma¨kinen et al.
Ganem LG, Trottier E, Anderson A, and Jefcoate CR (1999) Phenobarbital induction
Quack M and Carlberg C (2000) Ligand-triggered stabilization of vitamin D receptor/
of CYP2B1/2 in primary hepatocytes: endocrine regulation and evidence for a
retinoid X receptor heterodimer conformations on DR4-type response elements.
single pathway for multiple inducers. Toxicol Appl Pharmacol 155:32– 42.
J Mol Biol 296:743–756.
Goodwin B, Hodgson E, and Liddle C (1999) The orphan human pregnane X receptor
Quattrochi LC and Guzelian PS (2001) CYP3A regulation: from pharmacology to
mediates the transcriptional activation of CYP3A4 by rifampicin through a distal
nuclear receptors. Drug Metab Dispos 29:615– 622.
enhancer module. Mol Pharmacol 56:1329 –1339.
Schmiedlin-Ren P, Thummel KE, Fisher JM, Paine MF, Lown KS, and Watkins PB
Goodwin B, Moore LB, Stoltz CM, McKee DD, and Kliewer SA (2001) Regulation of
(1997) Expression of enzymatically active CYP3A4 by Caco-2 cells grown on
the human CYP2B6 gene by the nuclear pregnane X receptor. Mol Pharmacol
extracellular matrix-coated permeable supports in the presence of 1␣,25-
dihydroxyvitamin D3. Mol Pharmacol 51:741–754.
Harbers M, Wahlstro¨m GM, and Vennstro¨m B (1996) Transactivation by the thyroid
Schuetz EG, Strom S, Yasuda K, Lecureur V, Assem M, Brimer C, Lamba J, Kim RB,
hormone receptor is dependent on the spacer sequence in hormone response
Ramachandran V, Komoroski BJ, et al. (2001) Disrupted bile acid homeostasis
elements containing directly repeated half-sites. Nucl Acids Res 24:2252–2259.
reveals an unexpected interaction among nuclear hormone receptors, transporters
Honkakoski P, Jaaskelainen I, Kortelahti M, and Urtti A (2001) A novel drug-
and cytochrome P450. J Biol Chem 276:39411–39418.
regulated gene expression system based on the nuclear receptor constitutive
Sher T, Yi HF, McBride OW, and Gonzalez FJ (1993) cDNA cloning, chromosomal
androstane receptor (CAR). Pharm Res 18:146 –150.
mapping and functional characterization of the human peroxisome proliferator-
Honkakoski P, Moore R, Gynther J, and Negishi M (1996) Characterization of
activated receptor. Biochemistry 32:5598 –5604.
phenobarbital-inducible Cyp2b10 gene transcription in primary hepatocytes.
Smirlis D, Muangmoonchai R, Edwards M, Phillips IR, and Shephard EA (2001)
J Biol Chem 271:9746 –9753.
Orphan receptor promiscuity in the induction of cytochromes P450 by xenobiotics.
Honkakoski P and Negishi M (1998) Protein serine/threonine phosphatase inhibitors
J Biol Chem 276:12822–12826.
suppress phenobarbital-induced Cyp2b10 gene transcription in mouse primary
Smith G, Henderson CJ, Parker MG, White R, Bars RG, and Wolf CR (1993)
hepatocytes. Biochem J 330:889 – 895.
Honkakoski P and Negishi M (2000) Regulation of cytochrome P450 (CYP) genes by
1,4-Bis[2-(3,5-dichloropyridyloxy)]benzene, an extremely potent modulator of
nuclear receptors. Biochem J 347:321–337.
mouse hepatic cytochrome P-450 gene expression. Biochem J 289:807– 813.
Honkakoski P, Zelko I, Sueyoshi T, and Negishi M (1998) The orphan nuclear
Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, and Negishi M (1999) The re-
receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-
pressed nuclear receptor CAR responds to phenobarbital in activating the human
responsive enhancer module of the CYP2B gene. Mol Cell Biol 18:5652–5658.
CYP2B6 gene. J Biol Chem 274:6043– 6046.
Hu X and Lazar MA (1999) The CoRNR motif controls the recruitment of corepres-
Teboul M, Enmark E, Li Q, Wikstrom AC, Pelto-Huikko M and Gustafsson J-Å
sors by nuclear hormone receptors. Nature (Lond) 402:93–96.
(1995) OR-1, member of the nuclear receptor superfamily that interacts with the
Janowski BA, Willy PJ, Devi TR, Falck JR, and Mangelsdorf DJ (1996) An oxysterol
9-cis-retinoic acid receptor. Proc Natl Acad Sci USA 92:2096 –2100.
signalling pathway mediated by the nuclear receptor LXR␣. Nature (Lond) 383:
Tzameli I, Pissios P, Schuetz EG, and Moore DD (2000) The xenobiotic compound
1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear re-
Kahlert S, Nuedling S, van Eickels M, Vetter H, Meyer R, and Grohe C (2000)
ceptor CAR. Mol Cell Biol 20:2951–2958.
Estrogen receptor alpha rapidly activates the IGF-1 receptor pathway. J Biol
Voegel JJ, Heine MJ, Zechel C, Chambon P, and Gronemeyer H (1996) TIF2, a 160
kDa transcriptional mediator for the ligand-dependent activation function AF-2 of
Kim J, Min G, and Kemper B (2001) Chromatin assembly enhances binding to the
nuclear receptors. EMBO (Eur Mol Biol Organ) J 15:3667–3675.
CYP2B1 pheno- barbital-responsive unit (PBRU) of nuclear factor-1, which binds
Wan YY, An D, Cai Y, Repa JJ, Chen TH, Flores M, Postic C, Magnuson MA, Chen
simultaneously with constitutive androstane receptor (CAR)/retinoid X receptor
J, Chien KR, et al. (2000) Hepatocyte-specific mutation establishes retinoid X
(RXR) and enhances CAR/RXR-mediated activation of the PBRU. J Biol Chem
receptor ␣ as a heterodimeric integrator of multiple physiological processes in the
liver. Mol Cell Biol 20:4436 – 4444.
Laffitte BA, Kast HR, Nguyen CM, Zavacki AM, Moore DD, and Edwards PA (2000)
Waxman DJ (1999) P450 gene induction by structurally diverse xenochemicals:
Identification of the DNA binding specificity and potential target genes for the
central role of nuclear receptors CAR, PXR and PPAR. Arch Biochem Biophys
farnesoid X-activated receptor. J Biol Chem 275:10638 –10647.
Lehmann JM, McKee DD, Watson MA, Willson TM, Moore T, and Kliewer SA (1998)
Wei P, Zhang J, Egan-Hafley M, Liang S, and Moore DD (2000) The nuclear receptor
The human orphan nuclear receptor PXR is activated by compounds that regulate
CAR mediates specific xenobiotic induction of drug metabolism. Nature (Lond)
CYP3A4 gene expression and cause drug interactions. J Clin Invest 102:1016 –
Xie W, Barwick JL, Downes M, Blumberg B, Simon CM, Nelson MC, Neuschwander-
Mangelsdorf DJ and Evans RM (1995) The RXR heterodimers and orphan receptors.
Tetri BA, Brunt EM, Guzelian PS, and Evans RM (2000a) Humanized xenobiotic
Cell 83:841– 850.
response in mice expressing nuclear receptor SXR. Nature (Lond) 406:435– 438.
Moore LB, Parks DJ, Jones SA, Bledsoe RK, Consler TG, Stimmel J, Goodwin B,
Xie W, Barwick JL, Simon CM, Pierce AM, Safe S, Blumberg B, Guzelian PS, and
Liddle C, Blanchard SG, Willson TM, et al. (2000) Orphan nuclear receptors
Evans RM (2000b) Reciprocal activation of xenobiotic response genes by nuclear
constitutive androstane receptor and pregnane X receptor share xenobiotic and
receptors SXR/PXR and CAR. Genes Dev 14:3014 –3023.
steroid ligands. J Biol Chem 275:15122–15127.
Zelent A, Krust A, Petkovich M, Kastner P, and Chambon P (1989) Cloning of murine
Nishikawa J, Saito K, Goto J, Dakeyama F, Matsuo M, and Nishihara T (1999) New
alpha and beta retinoic acid receptors and a novel receptor gamma predominantly
screening methods for chemicals with hormonal activities using interaction of
expressed in skin. Nature (Lond) 339:714 –717.
nuclear hormone receptor with coactivator. Toxicol Appl Pharmacol 154:76 – 83.
Paquet Y, Trottier E, Beaudet MJ, and Anderson A (2000) Mutational analysis of the
Zhang J and Lazar MA (2000) The mechanism of action of thyroid hormones. Annu
CYP2B2 pheno- barbital response unit and inhibitory effect of the constitutive
Rev Physiol 62:439 – 466.
androstane receptor on phenobarbital responsiveness. J Biol Chem 275:38427–
38436.
Pellinen P, Honkakoski P, Stenback F, Niemitz M, Alhava E, Pelkonen O, Lang MA,
Address correspondence to: Dr. Paavo Honkakoski, Department of Phar-
and Pasanen M (1994) Cocaine N-demethylation and the metabolism-related hep-
maceutics, University of Kuopio, P.O.Box 1627, FIN-70211 Kuopio, Finland.
atotoxicity can be prevented by cytochrome P450 3A inhibitors. Eur J Pharmacol
Source: http://ita-suomenbiopankki.fi/documents/1076695/1076724/2002-4.pdf/03a0521f-d763-4420-a4c8-81519f2239b2
Final Report Snap ‘n Dose David Xue (Programmer) Niraj Mistry (Apper) Pooja Viswanathan (Programmer) Total Report Word Count (excluding title page & sample projects page): 2485 (Penalty - 0) Total Apper Context Word Count: 499 (Penalty - 0) Final Report Introduction Fever is the most common and concerning reason for which parents bring their children
William E. Seidelman MDScience and Inhumanity: The Kaiser-Wilhelm/Max Planck Society First Published in: If Not Now an e-journal Volume 2, Winter 2000 Revised February 18, 2001. One hundred years ago this past December a German scientist by the name of Max Karl Ernst Ludwig Planck gave a lecture in Berlin to the German Physical







